Mitochondrial genomes reveal slow rates of molecular evolution and the timing of speciation in beavers (Castor), one of the largest rodent species
- PMID: 21307956
- PMCID: PMC3030560
- DOI: 10.1371/journal.pone.0014622
Mitochondrial genomes reveal slow rates of molecular evolution and the timing of speciation in beavers (Castor), one of the largest rodent species
Abstract
Background: Beavers are one of the largest and ecologically most distinct rodent species. Little is known about their evolution and even their closest phylogenetic relatives have not yet been identified with certainty. Similarly, little is known about the timing of divergence events within the genus Castor.
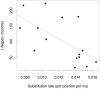
Methodology/principal findings: We sequenced complete mitochondrial genomes from both extant beaver species and used these sequences to place beavers in the phylogenetic tree of rodents and date their divergence from other rodents as well as the divergence events within the genus Castor. Our analyses support the phylogenetic position of beavers as a sister lineage to the scaly tailed squirrel Anomalurus within the mouse related clade. Molecular dating places the divergence time of the lineages leading to beavers and Anomalurus as early as around 54 million years ago (mya). The living beaver species, Castor canadensis from North America and Castor fiber from Eurasia, although similar in appearance, appear to have diverged from a common ancestor more than seven mya. This result is consistent with the hypothesis that a migration of Castor from Eurasia to North America as early as 7.5 mya could have initiated their speciation. We date the common ancestor of the extant Eurasian beaver relict populations to around 210,000 years ago, much earlier than previously thought. Finally, the substitution rate of Castor mitochondrial DNA is considerably lower than that of other rodents. We found evidence that this is correlated with the longer life span of beavers compared to other rodents.
Conclusions/significance: A phylogenetic analysis of mitochondrial genome sequences suggests a sister-group relationship between Castor and Anomalurus, and allows molecular dating of species divergence in congruence with paleontological data. The implementation of a relaxed molecular clock enabled us to estimate mitochondrial substitution rates and to evaluate the effect of life history traits on it.
Conflict of interest statement
Figures

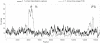


References
-
- Baker BW, Hill EP. Beaver (Castor canadensis). In: Feldhamer GA, Thompson BC, Chapman JA, editors. Wild Mammals of North America: biology, management, and conservation Baltimore, MD The Johns Hopkins University Press; 2003.
-
- Djoshkin WW, Safonow WG. Germany: Die Neue Brehm-Bücherei, A Ziemsen Verlag; 1972. Die Biber der alten und neuen Welt.
-
- Adkins RM, Walton AH, Honeycutt RL. Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Molecular Phylogenetics and Evolution. 2003;26:409–420. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

