Inferring cell cycle feedback regulation from gene expression data
- PMID: 21310265
- PMCID: PMC3143236
- DOI: 10.1016/j.jbi.2011.02.002
Inferring cell cycle feedback regulation from gene expression data
Abstract
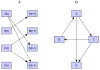
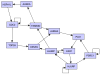
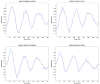
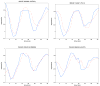
Feedback control is an important regulatory process in biological systems, which confers robustness against external and internal disturbances. Genes involved in feedback structures are therefore likely to have a major role in regulating cellular processes. Here we rely on a dynamic Bayesian network approach to identify feedback loops in cell cycle regulation. We analyzed the transcriptional profile of the cell cycle in HeLa cancer cells and identified a feedback loop structure composed of 10 genes. In silico analyses showed that these genes hold important roles in system's dynamics. The results of published experimental assays confirmed the central role of 8 of the identified feedback loop genes in cell cycle regulation. In conclusion, we provide a novel approach to identify critical genes for the dynamics of biological processes. This may lead to the identification of therapeutic targets in diseases that involve perturbations of these dynamics.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Threshold-dominated regulation hides genetic variation in gene expression networks.BMC Syst Biol. 2007 Dec 6;1:57. doi: 10.1186/1752-0509-1-57. BMC Syst Biol. 2007. PMID: 18062810 Free PMC article.
-
Reliability of transcriptional cycles and the yeast cell-cycle oscillator.PLoS Comput Biol. 2010 Jul 8;6(7):e1000842. doi: 10.1371/journal.pcbi.1000842. PLoS Comput Biol. 2010. PMID: 20628620 Free PMC article.
-
Reconstruction of gene regulatory modules in cancer cell cycle by multi-source data integration.PLoS One. 2010 Apr 21;5(4):e10268. doi: 10.1371/journal.pone.0010268. PLoS One. 2010. PMID: 20422009 Free PMC article.
-
Computational and experimental approaches for modeling gene regulatory networks.Curr Pharm Des. 2007;13(14):1415-36. doi: 10.2174/138161207780765945. Curr Pharm Des. 2007. PMID: 17504165 Review.
-
Inference of dynamic networks using time-course data.Brief Bioinform. 2014 Mar;15(2):212-28. doi: 10.1093/bib/bbt028. Epub 2013 May 21. Brief Bioinform. 2014. PMID: 23698724 Review.
Cited by
-
Application of a novel numerical simulation to biochemical reaction systems.Front Cell Dev Biol. 2024 Sep 6;12:1351974. doi: 10.3389/fcell.2024.1351974. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39310225 Free PMC article.
-
Marco Ramoni: an appreciation of academic achievement.J Am Med Inform Assoc. 2011 Jul-Aug;18(4):367-9. doi: 10.1136/amiajnl-2011-000218. Epub 2011 Apr 7. J Am Med Inform Assoc. 2011. PMID: 21474623 Free PMC article.
-
Recent development and biomedical applications of probabilistic Boolean networks.Cell Commun Signal. 2013 Jul 1;11:46. doi: 10.1186/1478-811X-11-46. Cell Commun Signal. 2013. PMID: 23815817 Free PMC article. Review.
References
-
- Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–37. - PubMed
-
- Csete ME, Doyle JC. Reverse engineering of biological complexity. Science. 2002;295:1664–9. - PubMed
-
- Thomas R, Thieffry D, Kaufman M. Dynamical behaviour of biological regulatory networks--I. Biological role of feedback loops and practical use of the concept of the loop-characteristic state. Bull Math Biol. 1995;57:247–276. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

