Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma
- PMID: 21316601
- PMCID: PMC3040414
- DOI: 10.1016/j.ccr.2010.12.023
Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma
Abstract
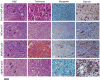
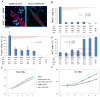
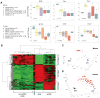
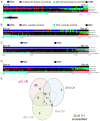
Embryonal rhabdomyosarcoma (eRMS) shows the most myodifferentiation among sarcomas, yet the precise cell of origin remains undefined. Using Ptch1, p53 and/or Rb1 conditional mouse models and controlling prenatal or postnatal myogenic cell of origin, we demonstrate that eRMS and undifferentiated pleomorphic sarcoma (UPS) lie in a continuum, with satellite cells predisposed to giving rise to UPS. Conversely, p53 loss in maturing myoblasts gives rise to eRMS, which have the highest myodifferentiation potential. Regardless of origin, Rb1 loss modifies tumor phenotype to mimic UPS. In human sarcomas that lack pathognomic chromosomal translocations, p53 loss of function is prevalent, whereas Shh or Rb1 alterations likely act primarily as modifiers. Thus, sarcoma phenotype is strongly influenced by cell of origin and mutational profile.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




Comment in
-
New insights into the origin and the genetic basis of rhabdomyosarcomas.Cancer Cell. 2011 Feb 15;19(2):157-9. doi: 10.1016/j.ccr.2011.01.044. Cancer Cell. 2011. PMID: 21316595 Free PMC article.
References
-
- Arndt CA, Hammond S, Rodeberg D, Qualman S. Significance of persistent mature rhabdomyoblasts in bladder/prostate rhabdomyosarcoma: Results from IRS IV. J Pediatr Hematol Oncol. 2006;28:563–567. - PubMed
-
- Barr FG, Smith LM, Lynch JC, Strzelecki D, Parham DM, Qualman SJ, Breitfeld PP. Examination of gene fusion status in archival samples of alveolar rhabdomyosarcoma entered on the Intergroup Rhabdomyosarcoma Study-III trial: a report from the Children's Oncology Group. J Mol Diagn. 2006;8:202–208. - PMC - PubMed
-
- Beddis IR, Mott MG, Bullimore J. Case report: nasopharyngeal rhabdomyosarcoma and Gorlin's naevoid basal cell carcinoma syndrome. Med Pediatr Oncol. 1983;11:178–179. - PubMed
-
- Breneman JC, Lyden E, Pappo AS, Link MP, Anderson JR, Parham DM, Qualman SJ, Wharam MD, Donaldson SS, Maurer HM, et al. Prognostic factors and clinical outcomes in children and adolescents with metastatic rhabdomyosarcoma--a report from the Intergroup Rhabdomyosarcoma Study IV. J Clin Oncol. 2003;21:78–84. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

