Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis
- PMID: 21317241
- PMCID: PMC3049287
- DOI: 10.1101/gad.2016311
Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis
Abstract
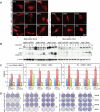
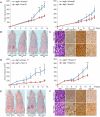
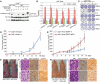
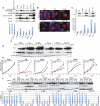
Autophagy is a catabolic pathway used by cells to support metabolism in response to starvation and to clear damaged proteins and organelles in response to stress. We report here that expression of a H-ras(V12) or K-ras(V12) oncogene up-regulates basal autophagy, which is required for tumor cell survival in starvation and in tumorigenesis. In Ras-expressing cells, defective autophagosome formation or cargo delivery causes accumulation of abnormal mitochondria and reduced oxygen consumption. Autophagy defects also lead to tricarboxylic acid (TCA) cycle metabolite and energy depletion in starvation. As mitochondria sustain viability of Ras-expressing cells in starvation, autophagy is required to maintain the pool of functional mitochondria necessary to support growth of Ras-driven tumors. Human cancer cell lines bearing activating mutations in Ras commonly have high levels of basal autophagy, and, in a subset of these, down-regulating the expression of essential autophagy proteins impaired cell growth. As cancers with Ras mutations have a poor prognosis, this "autophagy addiction" suggests that targeting autophagy and mitochondrial metabolism are valuable new approaches to treat these aggressive cancers.
Figures
Comment in
-
Interplay between autophagy and metabolism in Ras mutation-induced tumorigenesis.Asian J Androl. 2011 Jul;13(4):610-1. doi: 10.1038/aja.2011.31. Epub 2011 Apr 18. Asian J Androl. 2011. PMID: 21499280 Free PMC article. No abstract available.
-
Autophagy: limiting factors.Nat Rev Cancer. 2011 May;11(5):313. doi: 10.1038/nrc3054. Epub 2011 Apr 7. Nat Rev Cancer. 2011. PMID: 21508966 No abstract available.
References
-
- Degenhardt K, Chen G, Lindsten T, White E 2002a. BAX and BAK mediate p53-independent suppression of tumorigenesis. Cancer Cell 2: 193–203 - PubMed
-
- Degenhardt K, Sundararajan R, Lindsten T, Thompson C, White E 2002b. Bax and Bak independently promote cytochrome C release from mitochondria. J Biol Chem 277: 14127–14134 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
