Contribution of rapid evolution of the luxR-luxI intergenic region to the diverse bioluminescence outputs of Vibrio fischeri strains isolated from different environments
- PMID: 21317265
- PMCID: PMC3067414
- DOI: 10.1128/AEM.02643-10
Contribution of rapid evolution of the luxR-luxI intergenic region to the diverse bioluminescence outputs of Vibrio fischeri strains isolated from different environments
Abstract
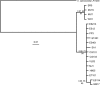
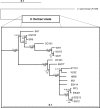
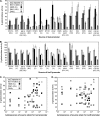
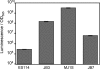
Vibrio fischeri serves as a valuable model of bacterial bioluminescence, its regulation, and its functional significance. Light output varies more than 10,000-fold in wild-type isolates from different environments, yet dim and bright strains have similar organization of the light-producing lux genes, with the activator-encoding luxR divergently transcribed from luxICDABEG. By comparing the genomes of bright strain MJ11 and the dimmer ES114, we found that the lux region has diverged more than most shared orthologs, including those flanking lux. Divergence was particularly high in the intergenic sequence between luxR and luxI. Analysis of the intergenic lux region from 18 V. fischeri strains revealed that, with one exception, sequence divergence essentially mirrored strain phylogeny but with relatively high substitution rates. The bases conserved among intergenic luxR-luxI sequences included binding sites for known regulators, such as LuxR and ArcA, and bases of unknown significance, including a striking palindromic repeat. By using this collection of diverse luxR-luxI regions, we found that expression of P(luxI)-lacZ but not P(luxR)-lacZ transcriptional reporters correlated with the luminescence output of the strains from which the promoters originated. We also found that exchange of a small stretch of the luxI-luxR intergenic region between two strains largely reversed their relative brightness. Our results show that the luxR-luxI intergenic region contributes significantly to the variable luminescence output among V. fischeri strains isolated from different environments, although other elements of strain backgrounds also contribute. Moreover, the lux system appears to have evolved relatively rapidly, suggesting unknown environment-specific selective pressures.
Figures



References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Ast, J. C., H. Urbanczyk, and P. V. Dunlap. 2009. Multi-gene analysis reveals previously unrecognized phylogenetic diversity in Aliivibrio. Syst. Appl. Microbiol. 32:379-386. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

