Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis
- PMID: 21317340
- PMCID: PMC3091125
- DOI: 10.1104/pp.110.169813
Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis
Abstract
Plant reproduction depends on the concerted activation of many genes to ensure correct communication between pollen and pistil. Here, we queried the whole transcriptome of Arabidopsis (Arabidopsis thaliana) in order to identify genes with specific reproductive functions. We used the Affymetrix ATH1 whole genome array to profile wild-type unpollinated pistils and unfertilized ovules. By comparing the expression profile of pistils at 0.5, 3.5, and 8.0 h after pollination and applying a number of statistical and bioinformatics criteria, we found 1,373 genes differentially regulated during pollen-pistil interactions. Robust clustering analysis grouped these genes in 16 time-course clusters representing distinct patterns of regulation. Coregulation within each cluster suggests the presence of distinct genetic pathways, which might be under the control of specific transcriptional regulators. A total of 78% of the regulated genes were expressed initially in unpollinated pistil and/or ovules, 15% were initially detected in the pollen data sets as enriched or preferentially expressed, and 7% were induced upon pollination. Among those, we found a particular enrichment for unknown transcripts predicted to encode secreted proteins or representing signaling and cell wall-related proteins, which may function by remodeling the extracellular matrix or as extracellular signaling molecules. A strict regulatory control in various metabolic pathways suggests that fine-tuning of the biochemical and physiological cellular environment is crucial for reproductive success. Our study provides a unique and detailed temporal and spatial gene expression profile of in vivo pollen-pistil interactions, providing a framework to better understand the basis of the molecular mechanisms operating during the reproductive process in higher plants.
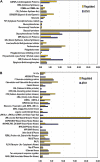
Figures





References
-
- Aloni R, Aloni E, Langhans M, Ullrich CI. (2006) Role of auxin in regulating Arabidopsis flower development. Planta 223: 315–328 - PubMed
-
- Boavida LC, Becker JD, Feijó JA. (2005a) The making of gametes in higher plants. Int J Dev Biol 49: 595–614 - PubMed
-
- Boavida LC, Vieira AM, Becker JD, Feijó JA. (2005b) Gametophyte interaction and sexual reproduction: how plants make a zygote. Int J Dev Biol 49: 615–632 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

