Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells
- PMID: 21317531
- PMCID: PMC3049389
- DOI: 10.1172/JCI44635
Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells
Abstract
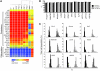
Human induced pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs) are promising candidate cell sources for regenerative medicine. However, despite the common ability of hiPSCs and hESCs to differentiate into all 3 germ layers, their functional equivalence at the single cell level remains to be demonstrated. Moreover, single cell heterogeneity amongst stem cell populations may underlie important cell fate decisions. Here, we used single cell analysis to resolve the gene expression profiles of 362 hiPSCs and hESCs for an array of 42 genes that characterize the pluripotent and differentiated states. Comparison between single hESCs and single hiPSCs revealed markedly more heterogeneity in gene expression levels in the hiPSCs, suggesting that hiPSCs occupy an alternate, less stable pluripotent state. hiPSCs also displayed slower growth kinetics and impaired directed differentiation as compared with hESCs. Our results suggest that caution should be exercised before assuming that hiPSCs occupy a pluripotent state equivalent to that of hESCs, particularly when producing differentiated cells for regenerative medicine aims.
Figures


References
-
- Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

