Structural basis for engagement by complement factor H of C3b on a self surface
- PMID: 21317894
- PMCID: PMC3512577
- DOI: 10.1038/nsmb.2018
Structural basis for engagement by complement factor H of C3b on a self surface
Abstract
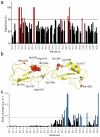
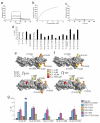
Complement factor H (FH) attenuates C3b molecules tethered by their thioester domains to self surfaces and thereby protects host tissues. Factor H is a cofactor for initial C3b proteolysis that ultimately yields a surface-attached fragment (C3d) corresponding to the thioester domain. We used NMR and X-ray crystallography to study the C3d-FH19-20 complex in atomic detail and identify glycosaminoglycan-binding residues in factor H module 20 of the C3d-FH19-20 complex. Mutagenesis justified the merging of the C3d-FH19-20 structure with an existing C3b-FH1-4 crystal structure. We concatenated the merged structure with the available FH6-8 crystal structure and new SAXS-derived FH1-4, FH8-15 and FH15-19 envelopes. The combined data are consistent with a bent-back factor H molecule that binds through its termini to two sites on one C3b molecule and simultaneously to adjacent polyanionic host-surface markers.
Figures




Comment in
-
In self-defense.Nat Struct Mol Biol. 2011 Apr;18(4):401-2. doi: 10.1038/nsmb.2036. Epub 2011 Feb 20. Nat Struct Mol Biol. 2011. PMID: 21336275 No abstract available.
References
-
- Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–6. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

