Lipopolysaccharide-induced miR-1224 negatively regulates tumour necrosis factor-α gene expression by modulating Sp1
- PMID: 21320120
- PMCID: PMC3088963
- DOI: 10.1111/j.1365-2567.2010.03374.x
Lipopolysaccharide-induced miR-1224 negatively regulates tumour necrosis factor-α gene expression by modulating Sp1
Abstract
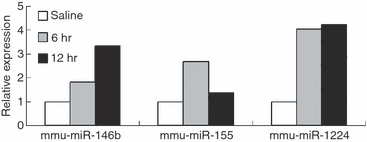
The innate immune response provides the initial defence mechanism against infection by other organisms. However, an excessive immune response will cause damage to host tissues. In an attempt to identify microRNAs (miRNAs) that regulate the innate immune response in inflammation and homeostasis, we examined the differential expression of miRNAs using microarray analysis in the spleens of mice injected intraperitoneally with lipopolysaccharide (LPS) and saline, respectively. Following challenge, we observed 19 miRNAs up-regulated (1.5-fold) in response to LPS. Among these miRNAs, miR-1224, whose expression level increased 5.7-fold 6 hr after LPS injection and 2.3-fold after 24 hr, was selected for further study. Tissue expression patterns showed that mouse miR-1224 is highly expressed in mouse spleen, kidney and lung. Transfection of miR-1224 mimics resulted in a decrease in basal tumour necrosis factor-α (TNF-α) promoter reporter gene activity and a down-regulation of LPS-induced TNF-α mRNA in RAW264.7 cells. With public databases of miRNA target prediction, miR-1224 was shown to bind to the 3' untranslated region (UTR) of Sp1 mRNA, whose coding product controls TNF-α expression at the transcriptional level. Furthermore, we found that in HEK-293 cells, the activity of the luciferase reporter bearing Sp1 mRNA 3' UTR was down-regulated significantly when transfected with miR-1224 mimics. After transfection of miR-1224 in RAW264.7 cells, nucleus Sp1 protein level decreased, and when endogenous miR-1224 was blocked, the decrease was abolished. Therefore, we initially speculated that miR-1224 was a negative regulator of TNF-α in an Sp1-dependent manner, which was confirmed in vivo by chromatin immunoprecipitation assay, and might be involved in regulating the LPS-mediated inflammatory responses.
© 2011 The Authors. Immunology © 2011 Blackwell Publishing Ltd.
Figures





Similar articles
-
MicroRNA-124 negatively regulates LPS-induced TNF-α production in mouse macrophages by decreasing protein stability.Acta Pharmacol Sin. 2016 Jul;37(7):889-97. doi: 10.1038/aps.2016.16. Epub 2016 Apr 11. Acta Pharmacol Sin. 2016. PMID: 27063215 Free PMC article.
-
Fascin regulates TLR4/PKC-mediated translational activation through miR-155 and miR-125b, which targets the 3' untranslated region of TNF-α mRNA.Immunol Invest. 2015;44(3):309-20. doi: 10.3109/08820139.2014.914533. Epub 2015 Apr 1. Immunol Invest. 2015. PMID: 25831081
-
MiR-34a inhibits lipopolysaccharide-induced inflammatory response through targeting Notch1 in murine macrophages.Exp Cell Res. 2012 Jun 10;318(10):1175-84. doi: 10.1016/j.yexcr.2012.03.018. Epub 2012 Mar 27. Exp Cell Res. 2012. PMID: 22483937
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
-
MicroRNAs: the fine-tuners of Toll-like receptor signalling.Nat Rev Immunol. 2011 Mar;11(3):163-75. doi: 10.1038/nri2957. Epub 2011 Feb 18. Nat Rev Immunol. 2011. PMID: 21331081 Review.
Cited by
-
Hsa_circ_0010957 knockdown attenuates lipopolysaccharide-induced HK2 cell injury by regulating the miR-1224-5p/IRAK1 axis.Cent Eur J Immunol. 2021;46(3):314-324. doi: 10.5114/ceji.2021.108772. Epub 2021 Sep 28. Cent Eur J Immunol. 2021. PMID: 34764803 Free PMC article.
-
miR‑494‑3p regulates lipopolysaccharide‑induced inflammatory responses in RAW264.7 cells by targeting PTEN.Mol Med Rep. 2019 May;19(5):4288-4296. doi: 10.3892/mmr.2019.10083. Epub 2019 Mar 26. Mol Med Rep. 2019. PMID: 30942409 Free PMC article.
-
Sp1 Plays an Important Role in Vascular Calcification Both In Vivo and In Vitro.J Am Heart Assoc. 2018 Mar 23;7(6):e007555. doi: 10.1161/JAHA.117.007555. J Am Heart Assoc. 2018. PMID: 29572322 Free PMC article.
-
Differentially Expressed miRNA-Gene Targets Related to Intramuscular Fat in Musculus Longissimus Dorsi of Charolais × Holstein F2-Crossbred Bulls.Genes (Basel). 2020 Jun 25;11(6):700. doi: 10.3390/genes11060700. Genes (Basel). 2020. PMID: 32630492 Free PMC article.
-
miR-1224 contributes to ischemic stroke-mediated natural killer cell dysfunction by targeting Sp1 signaling.J Neuroinflammation. 2021 Jun 12;18(1):133. doi: 10.1186/s12974-021-02181-4. J Neuroinflammation. 2021. PMID: 34118948 Free PMC article.
References
-
- Rasmussen SB, Reinert LS, Paludan SR. Innate recognition of intracellular pathogens: detection and activation of the first line of defense. Acta Pathol Microbiol Immunol Scand. 2009;117:323–37. - PubMed
-
- Us D. [Cytokine storm in avian influenza] Mikrobiyoloji bulteni. 2008;42:365–80. - PubMed
-
- Van Parijs L, Abbas AK. Homeostasis and self-tolerance in the immune system: turning lymphocytes off. Science (New York, NY) 1998;280:243–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

