An unusual tRNAThr derived from tRNAHis reassigns in yeast mitochondria the CUN codons to threonine
- PMID: 21321019
- PMCID: PMC3113583
- DOI: 10.1093/nar/gkr073
An unusual tRNAThr derived from tRNAHis reassigns in yeast mitochondria the CUN codons to threonine
Abstract
The standard genetic code is used by most living organisms, yet deviations have been observed in many genomes, suggesting that the genetic code has been evolving. In certain yeast mitochondria, CUN codons are reassigned from leucine to threonine, which requires an unusual tRNA(Thr) with an enlarged 8-nt anticodon loop ( ). To trace its evolutionary origin we performed a comprehensive phylogenetic analysis which revealed that evolved from yeast mitochondrial tRNA(His). To understand this tRNA identity change, we performed mutational and biochemical experiments. We show that Saccharomyces cerevisiae mitochondrial threonyl-tRNA synthetase (MST1) could attach threonine to both and the regular , but not to the wild-type tRNA(His). A loss of the first nucleotide (G(-1)) in tRNA(His) converts it to a substrate for MST1 with a K(m) value (0.7 μM) comparable to that of (0.3 μM), and addition of G(-1) to allows efficient histidylation by histidyl-tRNA synthetase. We also show that MST1 from Candida albicans, a yeast in which CUN codons remain assigned to leucine, could not threonylate , suggesting that MST1 has coevolved with . Our work provides the first clear example of a recent recoding event caused by alloacceptor tRNA gene recruitment.
Figures

 with an 8-nt anticodon loop and a UAG anticodon. (B)
with an 8-nt anticodon loop and a UAG anticodon. (B)  with a canonical UGU anticodon. (C) tRNAHis with a G−1 (circled, a major anti-determinant for ThrRS). The primary sequences of
with a canonical UGU anticodon. (C) tRNAHis with a G−1 (circled, a major anti-determinant for ThrRS). The primary sequences of  and tRNAHis are 72% identical (shaded).
and tRNAHis are 72% identical (shaded).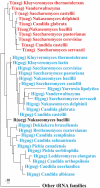
 and tRNAHis clusters are shown (marked red and blue, respectively), confirming monophyly of
and tRNAHis clusters are shown (marked red and blue, respectively), confirming monophyly of  and a sister group relationship to tRNAHis. The posterior probability support for the two tRNA groups is 1.0 (note that phylogenetic analysis with tRNA sequences depends on only few informative nucleotide positions, which does not allow to resolve the branching order within these groups). Removal of the anticodon sequence positions from the dataset did not change clustering into the two tRNA groups, nor did an analysis of the same datasets with maximum likelihood and the GTR model.
and a sister group relationship to tRNAHis. The posterior probability support for the two tRNA groups is 1.0 (note that phylogenetic analysis with tRNA sequences depends on only few informative nucleotide positions, which does not allow to resolve the branching order within these groups). Removal of the anticodon sequence positions from the dataset did not change clustering into the two tRNA groups, nor did an analysis of the same datasets with maximum likelihood and the GTR model.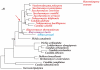
 (but not
(but not  ), and in these organisms CUN codons are assigned to Leu. The yeast species marked red, such as K. thermotolerans, have lost mitochondrial
), and in these organisms CUN codons are assigned to Leu. The yeast species marked red, such as K. thermotolerans, have lost mitochondrial  and obtained
and obtained  that decodes CUN codons as Thr. K. lactis is marked magenta as it has no CUN codons and no corresponding tRNA with a UAG anticodon. A. gossypii (marked blue) contains a tRNA species with a UAG anticodon, yet its identity of this tRNA and the amino acid reading CUN codons in A. gossypii remain obscure (to be discussed by BFL in Organelle Genetics: Evolution of Organelle Genomes and Gene Expression, Springer 2011).
that decodes CUN codons as Thr. K. lactis is marked magenta as it has no CUN codons and no corresponding tRNA with a UAG anticodon. A. gossypii (marked blue) contains a tRNA species with a UAG anticodon, yet its identity of this tRNA and the amino acid reading CUN codons in A. gossypii remain obscure (to be discussed by BFL in Organelle Genetics: Evolution of Organelle Genomes and Gene Expression, Springer 2011).
 (3 µM) and
(3 µM) and  (3 µM) by ScmtLeuRS (0.3 µM). (B) Threonylation of
(3 µM) by ScmtLeuRS (0.3 µM). (B) Threonylation of  (3 µM),
(3 µM),  (3 µM) and tRNAHis variants (3 µM) by ScMST1 (0.3 µM). (C) Histidylation of
(3 µM) and tRNAHis variants (3 µM) by ScMST1 (0.3 µM). (C) Histidylation of  (3 µM) and tRNAHis variants (3 µM) by ScmtHisRS (0.3 µM).
(3 µM) and tRNAHis variants (3 µM) by ScmtHisRS (0.3 µM).
 and tRNAHis. Three major differences between
and tRNAHis. Three major differences between  and tRNAHis sequences are indicated by boxes. Cc, Candida castellii; Cg, Candida glabrata; Kt, Kluyveromyces thermotolerans; Nb, Nakaseomyces bacillisporus; Nd, Nakaseomyces delphensis; Sca, Saccharomyces castellii; Sc, Saccharomyces cerevisiae; Sp, Saccharomyces pastorianus; Ss, Saccharomyces servazzii; Vp, Vanderwaltozyma polyspora.
and tRNAHis sequences are indicated by boxes. Cc, Candida castellii; Cg, Candida glabrata; Kt, Kluyveromyces thermotolerans; Nb, Nakaseomyces bacillisporus; Nd, Nakaseomyces delphensis; Sca, Saccharomyces castellii; Sc, Saccharomyces cerevisiae; Sp, Saccharomyces pastorianus; Ss, Saccharomyces servazzii; Vp, Vanderwaltozyma polyspora.
 and tRNAHis variants (3 µM) by C. albicans and S. pombe MST1 (0.3 µM). (A) CaMST1 and SpMST1 threonylate
and tRNAHis variants (3 µM) by C. albicans and S. pombe MST1 (0.3 µM). (A) CaMST1 and SpMST1 threonylate  but not
but not  . (B and C) CaMST1 and SpMST1 are unable to threonylate tRNAHis variants.
. (B and C) CaMST1 and SpMST1 are unable to threonylate tRNAHis variants.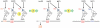
 , and
, and  was lost. (3) tRNAHis evolved to
was lost. (3) tRNAHis evolved to  , and CUN codons reemerged from various codons.
, and CUN codons reemerged from various codons.References
-
- Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. - PubMed
-
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. - PubMed
-
- Crick FH. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. - PubMed
-
- Moura GR, Paredes JA, Santos MA. Development of the genetic code: insights from a fungal codon reassignment. FEBS Lett. 2010;584:334–341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

