Xenopus HJURP and condensin II are required for CENP-A assembly
- PMID: 21321101
- PMCID: PMC3044122
- DOI: 10.1083/jcb.201005136
Xenopus HJURP and condensin II are required for CENP-A assembly
Erratum in
- J Cell Biol. 2011 Feb 21;192(4):preceding 569
- J Cell Biol. 2011 Mar 7;192(5):899
Abstract
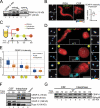
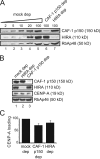
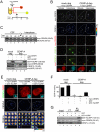
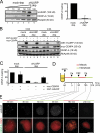
Centromeric protein A (CENP-A) is the epigenetic mark of centromeres. CENP-A replenishment is necessary in each cell cycle to compensate for the dilution associated to DNA replication, but how this is achieved mechanistically is largely unknown. We have developed an assay using Xenopus egg extracts that can recapitulate the spatial and temporal specificity of CENP-A deposition observed in human cells, providing us with a robust in vitro system amenable to molecular dissection. Here we show that this deposition depends on Xenopus Holliday junction-recognizing protein (xHJURP), a member of the HJURP/Scm3 family recently identified in yeast and human cells, further supporting the essential role of these chaperones in CENP-A loading. Despite little sequence homology, human HJURP can substitute for xHJURP. We also report that condensin II, but not condensin I, is required for CENP-A assembly and contributes to retention of centromeric CENP-A nucleosomes both in mitosis and interphase. We propose that the chromatin structure imposed by condensin II at centromeres enables CENP-A incorporation initiated by xHJURP.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

