Bisubstrate specificity in histidine/tryptophan biosynthesis isomerase from Mycobacterium tuberculosis by active site metamorphosis
- PMID: 21321225
- PMCID: PMC3048130
- DOI: 10.1073/pnas.1015996108
Bisubstrate specificity in histidine/tryptophan biosynthesis isomerase from Mycobacterium tuberculosis by active site metamorphosis
Abstract
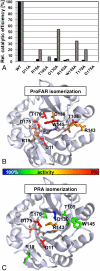
In histidine and tryptophan biosynthesis, two related isomerization reactions are generally catalyzed by two specific single-substrate enzymes (HisA and TrpF), sharing a similar (β/α)(8)-barrel scaffold. However, in some actinobacteria, one of the two encoding genes (trpF) is missing and the two reactions are instead catalyzed by one bisubstrate enzyme (PriA). To unravel the unknown mechanism of bisubstrate specificity, we used the Mycobacterium tuberculosis PriA enzyme as a model. Comparative structural analysis of the active site of the enzyme showed that PriA undergoes a reaction-specific and substrate-induced metamorphosis of the active site architecture, demonstrating its unique ability to essentially form two different substrate-specific actives sites. Furthermore, we found that one of the two catalytic residues in PriA, which are identical in both isomerization reactions, is recruited by a substrate-dependent mechanism into the active site to allow its involvement in catalysis. Comparison of the structural data from PriA with one of the two single-substrate enzymes (TrpF) revealed substantial differences in the active site architecture, suggesting independent evolution. To support these observations, we identified six small molecule compounds that inhibited both PriA-catalyzed isomerization reactions but had no effect on TrpF activity. Our data demonstrate an opportunity for organism-specific inhibition of enzymatic catalysis by taking advantage of the distinct ability for bisubstrate catalysis in the M. tuberculosis enzyme.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Gerlt JA, Raushel FM. Evolution of function in (β/α)8-barrel enzymes. Curr Opin Chem Biol. 2003;7:252–264. - PubMed
-
- Glasner ME, Gerlt JA, Babbitt PC. Mechanisms of protein evolution and their application to protein engineering. Adv Enzymol Relat Areas Mol Biol. 2007;75:193–239. - PubMed
-
- Khersonsky O, Roodveldt C, Tawfik DS. Enzyme promiscuity: Evolutionary and mechanistic aspects. Curr Opin Chem Biol. 2006;10:498–508. - PubMed
-
- Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. - PubMed
-
- Murzin AG. Biochemistry. Metamorphic proteins. Science. 2008;320:1725–1726. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

