An embryonic stage-specific enhancer within the murine β-globin locus mediates domain-wide histone hyperacetylation
- PMID: 21321362
- PMCID: PMC3109543
- DOI: 10.1182/blood-2010-08-302018
An embryonic stage-specific enhancer within the murine β-globin locus mediates domain-wide histone hyperacetylation
Abstract
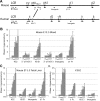
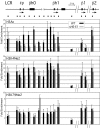
In mammalian nuclei, a select number of tissue-specific gene loci exhibit broadly distributed patterns of histone modifications, such as histone hyperacetylation, that are normally associated with active gene promoters. Previously, we characterized such hyperacetylated domains within mammalian β-globin gene loci, and determined that within the murine locus, neither the β-globin locus control region nor the gene promoters were required for domain formation. Here, we identify a developmentally specific erythroid enhancer, hypersensitive site-embryonic 1 (HS-E1), located within the embryonic β-globin domain in mouse, which is homologous to a region located downstream of the human embryonic ε-globin gene. This sequence exhibits nuclease hypersensitivity in primitive erythroid cells and acts as an enhancer in gain-of-function assays. Deletion of HS-E1 from the endogenous murine β-globin locus results in significant decrease in the expression of the embryonic β-globin genes and loss of the domain-wide pattern of histone hyperacetylation. The data suggest that HS-E1 is an enhancer that is uniquely required for β-like globin expression in primitive erythroid cells, and that it defines a novel class of enhancer that works in part by domain-wide modulation of chromatin structure.
Figures


References
-
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421(6921):448–453. - PubMed
-
- Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4(4):276–284. - PubMed
-
- Bulger M. Hyperacetylated chromatin domains: lessons from heterochromatin. J Biol Chem. 2005;280(23):21689–21692. - PubMed
-
- Fromm G, Bulger M. A spectrum of gene regulatory phenomena at mammalian beta-globin gene loci. Biochem Cell Biol. 2009;87(5):781–790. - PubMed
-
- Bender MA, Bulger M, Close J, Groudine M. Beta-globin gene switching and DNase I sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol Cell. 200;5(2):387–393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

