Advantages of tandem LC-MS for the rapid assessment of tissue-specific metabolic complexity using a pentafluorophenylpropyl stationary phase
- PMID: 21322650
- PMCID: PMC4158837
- DOI: 10.1021/pr1011119
Advantages of tandem LC-MS for the rapid assessment of tissue-specific metabolic complexity using a pentafluorophenylpropyl stationary phase
Abstract
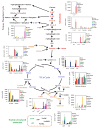
In this study, a tandem LC-MS (Waters Xevo TQ) MRM-based MS method was developed for rapid, broad profiling of hydrophilic metabolites from biological samples, in either positive or negative ion modes without the need for an ion pairing reagent, using a reversed-phase pentafluorophenylpropyl (PFPP) column. The developed method was successfully applied to analyze various biological samples from C57BL/6 mice, including urine, duodenum, liver, plasma, kidney, heart, and skeletal muscle. As result, a total 112 of hydrophilic metabolites were detected within 8 min of running time to obtain a metabolite profile of the biological samples. The analysis of this number of hydrophilic metabolites is significantly faster than previous studies. Classification separation for metabolites from different tissues was globally analyzed by PCA, PLS-DA and HCA biostatistical methods. Overall, most of the hydrophilic metabolites were found to have a "fingerprint" characteristic of tissue dependency. In general, a higher level of most metabolites was found in urine, duodenum, and kidney. Altogether, these results suggest that this method has potential application for targeted metabolomic analyzes of hydrophilic metabolites in a wide ranges of biological samples.
Figures




References
-
- Daviss B. Growing pains for metabolomics. The Scientist. 2005;19:25–28.
-
- Robertson D, Lindon JC, Nicholson JK, Holmes E, editors. Metabonomics in Toxicity Assessment. CRC Press; Boca Raton: 2005.
-
- Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, Bethell HWL, Clarke S, Schofield PM, McKilligin E, Mosedale D, Grainger DJ. Rapid and non-invasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR based metabonomics. Nature Med. 2002;8:1439–1445. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

