Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression
- PMID: 21324878
- PMCID: PMC3083081
- DOI: 10.1101/gr.097378.109
Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression
Erratum in
- Genome Res. 2011 Jun;21(6):999
Abstract
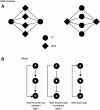
In any given cell, thousands of genes are expressed and work in concert to ensure the cell's function, fitness, and survival. Each gene, in turn, must be expressed at the proper time and in the proper amounts to ensure the appropriate functional outcome. The regulation and expression of some genes are highly robust; their expression is controlled by invariable expression programs. For instance, developmental gene expression is extremely similar in a given cell type from one individual to another. The expression of other genes is more variable: Their levels are noisy and are different from cell to cell and from individual to individual. This can be highly beneficial in physiological responses to outside cues and stresses. Recent advances have enabled the analysis of differential gene expression at a systems level. Gene regulatory networks (GRNs) involving interactions between large numbers of genes and their regulators have been mapped onto graphic diagrams that are used to visualize the regulatory relationships. The further characterization of GRNs has already uncovered global principles of gene regulation. Together with synthetic network biology, such studies are starting to provide insights into the transcriptional mechanisms that cause robust versus stochastic gene expression and their relationships to phenotypic robustness and variability. Here, we discuss GRNs and their topological properties in relation to transcriptional and phenotypic outputs in development and organismal physiology.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
