Subunit dimers of alpha-hemolysin expand the engineering toolbox for protein nanopores
- PMID: 21324910
- PMCID: PMC3077633
- DOI: 10.1074/jbc.M111.218164
Subunit dimers of alpha-hemolysin expand the engineering toolbox for protein nanopores
Abstract
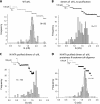
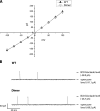
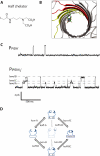
Staphylococcal α-hemolysin (αHL) forms a heptameric pore that features a 14-stranded transmembrane β-barrel. We attempted to force the αHL pore to adopt novel stoichiometries by oligomerizing subunit dimers generated by in vitro transcription and translation of a tandem gene. However, in vitro transcription and translation also produced truncated proteins, monomers, that were preferentially incorporated into oligomers. These oligomers were shown to be functional heptamers by single-channel recording and had a similar mobility to wild-type heptamers in SDS-polyacrylamide gels. Purified full-length subunit dimers were then prepared by using His-tagged protein. Again, single-channel recording showed that oligomers made from these dimers are functional heptamers, implying that one or more subunits are excluded from the central pore. Therefore, the αHL pore resists all structures except those that possess seven subunits immediately surrounding the central axis. Although we were not able to change the stoichiometry of the central pore of αHL by the concatenation of subunits, we extended our findings to prepare pores containing one subunit dimer and five monomers and purified them by SDS-PAGE. Two half-chelating ligands were then installed at adjacent sites, one on each subunit of the dimer. Single-channel recording showed that pores formed from this construct formed complexes with divalent metal ions in a similar fashion to pores containing two half-chelating ligands on the same subunit, confirming that the oligomers had assembled with seven subunits around the central lumen. The ability to incorporate subunit dimers into αHL pores increases the range of structures that can be obtained from engineered protein nanopores.
Figures

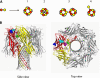





Similar articles
-
Ion Mobility-Mass Spectrometry Reveals That α-Hemolysin from Staphylococcus aureus Simultaneously Forms Hexameric and Heptameric Complexes in Detergent Micelle Solutions.Anal Chem. 2019 Aug 6;91(15):10204-10211. doi: 10.1021/acs.analchem.9b02243. Epub 2019 Jul 18. Anal Chem. 2019. PMID: 31282652 Free PMC article.
-
The internal cavity of the staphylococcal alpha-hemolysin pore accommodates approximately 175 exogenous amino acid residues.Biochemistry. 2005 Jun 28;44(25):8919-29. doi: 10.1021/bi0473713. Biochemistry. 2005. PMID: 15966717
-
A functional protein pore with a "retro" transmembrane domain.Protein Sci. 1999 Jun;8(6):1257-67. doi: 10.1110/ps.8.6.1257. Protein Sci. 1999. PMID: 10386875 Free PMC article.
-
Mode of action of beta-barrel pore-forming toxins of the staphylococcal alpha-hemolysin family.Toxicon. 2001 Nov;39(11):1661-72. doi: 10.1016/s0041-0101(01)00153-2. Toxicon. 2001. PMID: 11595629 Review.
-
Ion channels and bacterial infection: the case of beta-barrel pore-forming protein toxins of Staphylococcus aureus.FEBS Lett. 2003 Sep 18;552(1):54-60. doi: 10.1016/s0014-5793(03)00850-0. FEBS Lett. 2003. PMID: 12972152 Review.
Cited by
-
Ion Mobility-Mass Spectrometry Reveals That α-Hemolysin from Staphylococcus aureus Simultaneously Forms Hexameric and Heptameric Complexes in Detergent Micelle Solutions.Anal Chem. 2019 Aug 6;91(15):10204-10211. doi: 10.1021/acs.analchem.9b02243. Epub 2019 Jul 18. Anal Chem. 2019. PMID: 31282652 Free PMC article.
-
MspA nanopores from subunit dimers.PLoS One. 2012;7(6):e38726. doi: 10.1371/journal.pone.0038726. Epub 2012 Jun 18. PLoS One. 2012. PMID: 22719928 Free PMC article.
-
Engineered transmembrane pores.Curr Opin Chem Biol. 2016 Oct;34:117-126. doi: 10.1016/j.cbpa.2016.08.005. Epub 2016 Sep 20. Curr Opin Chem Biol. 2016. PMID: 27658267 Free PMC article. Review.
-
Detection of 3'-end RNA uridylation with a protein nanopore.ACS Nano. 2014 Feb 25;8(2):1364-74. doi: 10.1021/nn4050479. Epub 2013 Dec 31. ACS Nano. 2014. PMID: 24369707 Free PMC article.
-
Obstructing toxin pathways by targeted pore blockage.Chem Rev. 2012 Dec 12;112(12):6388-430. doi: 10.1021/cr300141q. Epub 2012 Oct 11. Chem Rev. 2012. PMID: 23057504 Free PMC article. Review. No abstract available.
References
-
- Howorka S., Siwy Z. (2009) Chem. Soc. Rev. 38, 2360–2384 - PubMed
-
- Siwy Z. S., Howorka S. (2010) Chem. Soc. Rev. 39, 1115–1132 - PubMed
-
- Panchal R. G., Cusack E., Cheley S., Bayley H. (1996) Nat. Biotechnol. 14, 852–856 - PubMed
-
- Eroglu A., Russo M. J., Bieganski R., Fowler A., Cheley S., Bayley H., Toner M. (2000) Nat. Biotechnol. 18, 163–167 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

