The population dynamics of bacteria in physically structured habitats and the adaptive virtue of random motility
- PMID: 21325053
- PMCID: PMC3053974
- DOI: 10.1073/pnas.1013499108
The population dynamics of bacteria in physically structured habitats and the adaptive virtue of random motility
Abstract
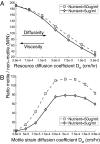
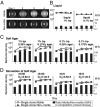
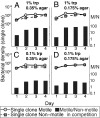
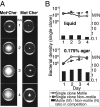
Why is motility so common in bacteria? An obvious answer to this ecological and evolutionary question is that in almost all habitats, bacteria need to go someplace and particularly in the direction of food. Although the machinery required for motility and chemotaxis (acquiring and processing the information needed to direct movement toward nutrients) are functionally coupled in contemporary bacteria, they are coded for by different sets of genes. Moreover, information that resources are more abundant elsewhere in a habitat would be of no value to a bacterium unless it already had the means to get there. Thus, motility must have evolved before chemotaxis, and bacteria with flagella and other machinery for propulsion in random directions must have an advantage over bacteria relegated to moving at the whim of external forces alone. However, what are the selection pressures responsible for the evolution and maintenance of undirected motility in bacteria? Here we use a combination of mathematical modeling and experiments with Escherichia coli to generate and test a parsimonious and ecologically general hypothesis for the existence of undirected motility in bacteria: it enables bacteria to move away from each other and thereby obtain greater individual shares of resources in physically structured environments. The results of our experiments not only support this hypothesis, but are quantitatively and qualitatively consistent with the predictions of our model.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Dinsdale EA, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. - PubMed
-
- NCBI-NIH . NCBI Prokaryotic Genomes. Bethesda: National Center for Biotechnology Information, National Institutes of Health; 2010.
-
- Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

