Testing for ancient admixture between closely related populations
- PMID: 21325092
- PMCID: PMC3144383
- DOI: 10.1093/molbev/msr048
Testing for ancient admixture between closely related populations
Abstract
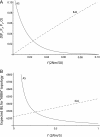
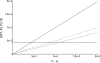
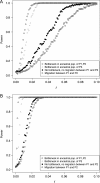
One enduring question in evolutionary biology is the extent of archaic admixture in the genomes of present-day populations. In this paper, we present a test for ancient admixture that exploits the asymmetry in the frequencies of the two nonconcordant gene trees in a three-population tree. This test was first applied to detect interbreeding between Neandertals and modern humans. We derive the analytic expectation of a test statistic, called the D statistic, which is sensitive to asymmetry under alternative demographic scenarios. We show that the D statistic is insensitive to some demographic assumptions such as ancestral population sizes and requires only the assumption that the ancestral populations were randomly mating. An important aspect of D statistics is that they can be used to detect archaic admixture even when no archaic sample is available. We explore the effect of sequencing error on the false-positive rate of the test for admixture, and we show how to estimate the proportion of archaic ancestry in the genomes of present-day populations. We also investigate a model of subdivision in ancestral populations that can result in D statistics that indicate recent admixture.
Figures



References
-
- Barton N. The role of hybridization in evolution. Mol Ecol. 2001;10:551–568. - PubMed
-
- Barton N, Hewitt G. Analysis of hybrid zones. Annu Rev Ecol Syst. 1985;16:113–148.
-
- Currat M, Ruedi M, Petit RJ, Excoffier L. The hidden side of invasions: massive introgression by local genes. Evolution. 2008;62(8):1908–1920. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

