Phylogenetic relationships among arecoid palms (Arecaceae: Arecoideae)
- PMID: 21325340
- PMCID: PMC3219489
- DOI: 10.1093/aob/mcr020
Phylogenetic relationships among arecoid palms (Arecaceae: Arecoideae)
Abstract
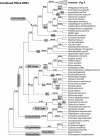
Background and aims: The Arecoideae is the largest and most diverse of the five subfamilies of palms (Arecaceae/Palmae), containing >50 % of the species in the family. Despite its importance, phylogenetic relationships among Arecoideae are poorly understood. Here the most densely sampled phylogenetic analysis of Arecoideae available to date is presented. The results are used to test the current classification of the subfamily and to identify priority areas for future research.
Methods: DNA sequence data for the low-copy nuclear genes PRK and RPB2 were collected from 190 palm species, covering 103 (96 %) genera of Arecoideae. The data were analysed using the parsimony ratchet, maximum likelihood, and both likelihood and parsimony bootstrapping.
Key results and conclusions: Despite the recovery of paralogues and pseudogenes in a small number of taxa, PRK and RPB2 were both highly informative, producing well-resolved phylogenetic trees with many nodes well supported by bootstrap analyses. Simultaneous analyses of the combined data sets provided additional resolution and support. Two areas of incongruence between PRK and RPB2 were strongly supported by the bootstrap relating to the placement of tribes Chamaedoreeae, Iriarteeae and Reinhardtieae; the causes of this incongruence remain uncertain. The current classification within Arecoideae was strongly supported by the present data. Of the 14 tribes and 14 sub-tribes in the classification, only five sub-tribes from tribe Areceae (Basseliniinae, Linospadicinae, Oncospermatinae, Rhopalostylidinae and Verschaffeltiinae) failed to receive support. Three major higher level clades were strongly supported: (1) the RRC clade (Roystoneeae, Reinhardtieae and Cocoseae), (2) the POS clade (Podococceae, Oranieae and Sclerospermeae) and (3) the core arecoid clade (Areceae, Euterpeae, Geonomateae, Leopoldinieae, Manicarieae and Pelagodoxeae). However, new data sources are required to elucidate ambiguities that remain in phylogenetic relationships among and within the major groups of Arecoideae, as well as within the Areceae, the largest tribe in the palm family.
Figures




References
-
- Asmussen CB, Chase MW. Coding and noncoding plastid DNA in palm systematics. American Journal of Botany. 2001;88:1103–1117. - PubMed
-
- Asmussen CB, Baker WJ, Dransfield J. Phylogeny of the palm family (Arecaceae) based on rps16 intron and trnL–trnF plastid DNA sequences. In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Melbourne: CSIRO; 2000. pp. 525–535.
-
- Asmussen CB, Dransfield J, Deickmann V, Barfod AS, Pintaud JC, Baker WJ. A new subfamily classification of the palm family (Arecaceae): evidence from plastid DNA phylogeny. Botanical Journal of the Linnean Society. 2006;151:15–38.
-
- Bacon CD, Feltus FA, Paterson AH, Bailey CD. Novel nuclear intron-spanning primers for Arecaceae evolutionary biology. Molecular Ecology Resources. 2008;8:211–214. - PubMed
-
- Barker FK, Lutzoni FM. The utility of the incongruence length difference test. Systematic Biology. 2002;51:625–637. - PubMed

