The Arabidopsis peptide kiss of death is an inducer of programmed cell death
- PMID: 21326210
- PMCID: PMC3061025
- DOI: 10.1038/emboj.2011.14
The Arabidopsis peptide kiss of death is an inducer of programmed cell death
Abstract
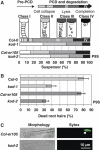
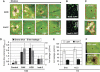
Programmed cell death (PCD) has a key role in defence and development of all multicellular organisms. In plants, there is a large gap in our knowledge of the molecular machinery involved at the various stages of PCD, especially the early steps. Here, we identify kiss of death (KOD) encoding a 25-amino-acid peptide that activates a PCD pathway in Arabidopsis thaliana. Two mutant alleles of KOD exhibited a reduced PCD of the suspensor, a single file of cells that support embryo development, and a reduced PCD of root hairs after a 55°C heat shock. KOD expression was found to be inducible by biotic and abiotic stresses. Furthermore, KOD expression was sufficient to cause death in leaves or seedlings and to activate caspase-like activities. In addition, KOD-induced PCD required light in leaves and was repressed by the PCD-suppressor genes AtBax inhibitor 1 and p35. KOD expression resulted in depolarization of the mitochondrial membrane, placing KOD above mitochondria dysfunction, an early step in plant PCD. A KOD∷GFP fusion, however, localized in the cytosol of cells and not mitochondria.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 - PubMed
-
- Bonneau L, Ge Y, Drury GE, Gallois P (2008) What happened to plant caspases? J Exp Bot 59: 491–499 - PubMed
-
- Bozhkov P, Filonova L, Suarez M (2005) Programmed cell death in plant embryogenesis. Curr Topics Dev Biol 67: 135–179 - PubMed
-
- Bozhkov PV, Filonova LH, Suarez MF, Helmersson A, Smertenko AP, Zhivotovsky B, von Arnold S (2003) VEIDase is a principal caspase-like activity involved in plant programmed cell death and essential for embryonic pattern formation. Cell Death Differ 11: 175–182 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

