Optimization based tumor classification from microarray gene expression data
- PMID: 21326602
- PMCID: PMC3033885
- DOI: 10.1371/journal.pone.0014579
Optimization based tumor classification from microarray gene expression data
Abstract
Background: An important use of data obtained from microarray measurements is the classification of tumor types with respect to genes that are either up or down regulated in specific cancer types. A number of algorithms have been proposed to obtain such classifications. These algorithms usually require parameter optimization to obtain accurate results depending on the type of data. Additionally, it is highly critical to find an optimal set of markers among those up or down regulated genes that can be clinically utilized to build assays for the diagnosis or to follow progression of specific cancer types. In this paper, we employ a mixed integer programming based classification algorithm named hyper-box enclosure method (HBE) for the classification of some cancer types with a minimal set of predictor genes. This optimization based method which is a user friendly and efficient classifier may allow the clinicians to diagnose and follow progression of certain cancer types.
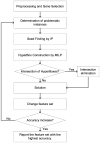
Methodology/principal findings: We apply HBE algorithm to some well known data sets such as leukemia, prostate cancer, diffuse large B-cell lymphoma (DLBCL), small round blue cell tumors (SRBCT) to find some predictor genes that can be utilized for diagnosis and prognosis in a robust manner with a high accuracy. Our approach does not require any modification or parameter optimization for each data set. Additionally, information gain attribute evaluator, relief attribute evaluator and correlation-based feature selection methods are employed for the gene selection. The results are compared with those from other studies and biological roles of selected genes in corresponding cancer type are described.
Conclusions/significance: The performance of our algorithm overall was better than the other algorithms reported in the literature and classifiers found in WEKA data-mining package. Since it does not require a parameter optimization and it performs consistently very high prediction rate on different type of data sets, HBE method is an effective and consistent tool for cancer type prediction with a small number of gene markers.
Conflict of interest statement
Figures


References
-
- Slonim DK. From patterns to pathways: gene expression data analysis comes of age. Nat Genet. 2002;32:502–508. - PubMed
-
- Schwarz G. Estimating the dimension of a model. Ann Statist. 1976;6:461–464.
-
- Kohavi G, John R. Wrappers for feature subset selection. Artif Intell. 1997;97:273–324.
-
- Wang Y, Tetko IV, Hall MA. Gene selection from microarray data for cancer classification a machine learning approach. Comp Biol Chem. 2005;29:37–46. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

