Two-component system RgfA/C activates the fbsB gene encoding major fibrinogen-binding protein in highly virulent CC17 clone group B Streptococcus
- PMID: 21326613
- PMCID: PMC3033900
- DOI: 10.1371/journal.pone.0014658
Two-component system RgfA/C activates the fbsB gene encoding major fibrinogen-binding protein in highly virulent CC17 clone group B Streptococcus
Abstract
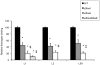
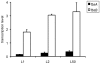
Group B streptococcus (GBS) strains with the highest ability to bind to human fibrinogen belong to the highly invasive clonal complex (CC) 17. To investigate the fibrinogen-binding mechanisms of CC17 strains, we determined the prevalence of fibrinogen-binding genes (fbsA and fbsB), and fbs regulator genes (rogB encoding an fbsA activator, rovS encoding an fbsA repressor and rgf encoding a two-component system [TCS] whose role on fbs genes was not determined yet) in a collection of 134 strains representing the major CCs of the species. We showed that specific gene combinations were related to particular CCs; only CC17 strains contained the fbsA, fbsB, and rgf genes combination. Non polar rgfAC deletion mutants of three CC17 serotype III strains were constructed. They showed a 3.2- to 5.1-fold increase of fbsA transcripts, a 4.8- to 6.7-fold decrease of fbsB transcripts, and a 52% to 68% decreased fibrinogen-binding ability, demonstrating that the RgfA/RgfC TCS inhibits the fbsA gene and activates the fbsB gene. The relative contribution of the two fbs genes in fibrinogen-binding ability was determined by constructing isogenic fbsA, fbsB, deletion mutants of the three CC17 strains. The ability to bind to fibrinogen was reduced by 49% to 57% in ΔfbsA mutants, and by 78% to 80% in ΔfbsB mutants, suggesting that FbsB protein plays a greater role in the fibrinogen-binding ability of CC17 strains. Moreover, the relative transcription level of fbsB gene was 9.2- to 12.7-fold higher than that of fbsA gene for the three wild type strains. Fibrinogen-binding ability could be restored by plasmid-mediated expression of rgfAC, fbsA, and fbsB genes in the corresponding deletion mutants. Thus, our results demonstrate that a specific combination of fbs genes and fbs regulator genes account for the high fibrinogen-binding ability of CC17 strains that may participate to their enhanced invasiveness for neonates as compared to strains of other CCs.
Conflict of interest statement
Figures
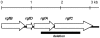


References
-
- Farley MM. Group B streptococcal disease in nonpregnant adults. Clin Infect Dis. 2001;33:556–561. - PubMed
-
- Bisharat N, Jones N, Marchaim D, Block C, Harding RM, et al. Population structure of group B streptococcus from a low-incidence region for invasive neonatal disease. Microbiology. 2005;151:1875–1881. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

