Molecular basis of transcription initiation in Archaea
- PMID: 21326901
- PMCID: PMC3023638
- DOI: 10.4161/trns.1.2.13189
Molecular basis of transcription initiation in Archaea
Abstract
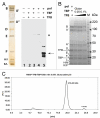
Compared with eukaryotes, the archaeal transcription initiation machinery-commonly known as the Pre-Initiation Complex-is relatively simple. The archaeal PIC consists of the TFIIB ortholog TFB, TBP, and an 11-subunit RNA polymerase (RNAP). The relatively small size of the entire archaeal PIC makes it amenable to structural analysis. Using purified RNAP, TFB, and TBP from the thermophile Pyrococcus furiosus, we assembled the biochemically active PIC at 65ºC. The intact archaeal PIC was isolated by implementing a cross-linking technique followed by size-exclusion chromatography, and the structure of this 440 kDa assembly was determined using electron microscopy and single-particle reconstruction techniques. Combining difference maps with crystal structure docking of various sub-domains, TBP and TFB were localized within the macromolecular PIC. TBP/TFB assemble near the large RpoB subunit and the RpoD/L "foot" domain behind the RNAP central cleft. This location mimics that of yeast TBP and TFIIB in complex with yeast RNAP II. Collectively, these results define the structural organization of the archaeal transcription machinery and suggest a conserved core PIC architecture.
Keywords: RNAP; archaea; cryo-electron microscopy; initiation; molecular fitting; transcription.
Figures




References
-
- Ebright RH. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol. 2000;304:687–698. - PubMed
-
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. - PubMed
-
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. - PubMed
-
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
