Folding kinetics of the cooperatively folded subdomain of the IκBα ankyrin repeat domain
- PMID: 21329696
- PMCID: PMC3081522
- DOI: 10.1016/j.jmb.2011.02.021
Folding kinetics of the cooperatively folded subdomain of the IκBα ankyrin repeat domain
Abstract
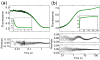
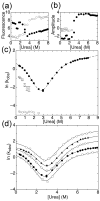
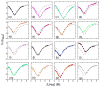
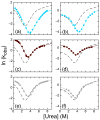
The ankyrin repeat (AR) domain of IκBα consists of a cooperative folding unit of roughly four ARs (AR1-AR4) and of two weakly folded repeats (AR5 and AR6). The kinetic folding mechanism of the cooperative subdomain, IκBα(67-206), was analyzed using rapid mixing techniques. Despite its apparent architectural simplicity, IκBα(67-206) displays complex folding kinetics, with two sequential on-pathway high-energy intermediates. The effect of mutations to or away from the consensus sequences of ARs on folding behavior was analyzed, particularly the GXTPLHLA motif, which have not been examined in detail previously. Mutations toward the consensus generally resulted in an increase in folding stability, whereas mutations away from the consensus resulted in decreased overall stability. We determined the free energy change upon mutation for three sequential transition state ensembles along the folding route for 16 mutants. We show that folding initiates with the formation of the interface of the outer helices of AR3 and AR4, and then proceeds to consolidate structure in these repeats. Subsequently, AR1 and AR2 fold in a concerted way in a single kinetic step. We show that this mechanism is robust to the presence of AR5 and AR6 as they do not strongly affect the folding kinetics. Overall, the protein appears to fold on a rather smooth energy landscape, where the folding mechanism conforms a one-dimensional approximation. However, we note that the AR does not necessarily act as a single folding element.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures



References
-
- Baeuerle PA, Baltimore D. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 1988;242:540–6. - PubMed
-
- Ferreiro DU, Cho SS, Komives EA, Wolynes PG. The energy landscape of modular repeat proteins: topology determines folding mechanism in the ankyrin family. J Mol Biol. 2005;354:679–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

