Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis
- PMID: 21330376
- PMCID: PMC3069410
- DOI: 10.1074/jbc.M110.186882
Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis
Abstract
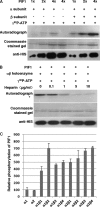
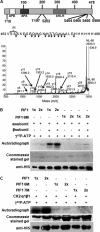
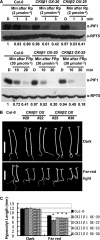
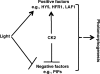
The phytochrome family of sensory photoreceptors interacts with phytochrome interacting factors (PIFs), repressors of photomorphogenesis, in response to environmental light signals and induces rapid phosphorylation and degradation of PIFs to promote photomorphogenesis. However, the kinase that phosphorylates PIFs is still unknown. Here we show that CK2 directly phosphorylates PIF1 at multiple sites. α1 and α2 subunits individually phosphorylated PIF1 weakly in vitro. However, each of four β subunits strongly stimulated phosphorylation of PIF1 by α1 or α2. Mapping of the phosphorylation sites identified seven Ser/Thr residues scattered throughout PIF1. Ser/Thr to Ala scanning mutations at all seven sites eliminated CK2-mediated phosphorylation of PIF1 in vitro. Moreover, the rate of degradation of the Ser/Thr to Ala mutant PIF1 was significantly reduced compared with wild-type PIF1 in transgenic plants. In addition, hypocotyl lengths of the mutant PIF1 transgenic plants were much longer than the wild-type PIF1 transgenic plants under light, suggesting that the mutant PIF1 is suppressing photomorphogenesis. Taken together, these data suggest that CK2-mediated phosphorylation enhances the light-induced degradation of PIF1 to promote photomorphogenesis.
Figures


References
-
- Bae G., Choi G. (2008) Annu. Rev. Plant Biol. 59, 281–311 - PubMed
-
- Schaefer E., Nagy F. (2006) Photomorphogenesis in Plants and Bacteria, 3rd Ed., Springer, Dordrecht, The Netherlands
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

