AFAP1L1 is a novel adaptor protein of the AFAP family that interacts with cortactin and localizes to invadosomes
- PMID: 21333378
- PMCID: PMC3085893
- DOI: 10.1016/j.ejcb.2010.11.016
AFAP1L1 is a novel adaptor protein of the AFAP family that interacts with cortactin and localizes to invadosomes
Abstract
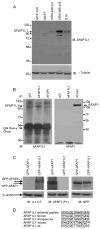
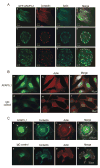
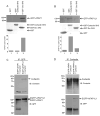
The actin-filament associated protein (AFAP) family of adaptor proteins consists of three members: AFAP1, AFAP1L1, and AFAP1L2/XB130 with AFAP1 being the best described as a cSrc binding partner and actin cross-linking protein. A homology search of AFAP1 recently identified AFAP1L1 which has a similar sequence, domain structure and cellular localization; however, based upon sequence variations, AFAP1L1 is hypothesized to have unique functions that are distinct from AFAP1. While AFAP1 has the ability to bind to the SH3 domain of the nonreceptor tyrosine kinase cSrc via an N-terminal SH3 binding motif, it was unable to bind cortactin. However, the SH3 binding motif of AFAP1L1 was more efficient at interacting with the SH3 domain of cortactin and not cSrc. AFAP1L1 was shown by fluorescence microscopy to decorate actin filaments and move to punctate actin structures and colocalize with cortactin, consistent with localization to invadosomes. Upon overexpression in A7r5 cells, AFAP1L1 had the ability to induce podosome formation and move to podosomes without stimulation. Immunohistochemical analysis of AFAP1L1 in human tissues shows differential expression when contrasted with AFAP1 with localization of AFAP1L1 to unique sites in muscle and the dentate nucleus of the brain where AFAP1 was not detectable. We hypothesize AFAP1L1 may play a similar role to AFAP1 in affecting changes in actin filaments and bridging interactions with binding partners, but we hypothesize that AFAP1L1 may forge unique protein interactions in which AFAP1 is less efficient, and these interactions may allow AFAP1L1 to affect invadosome formation.
Copyright © 2011 Elsevier GmbH. All rights reserved.
Figures




References
-
- Baisden JM, Gatesman AS, Cherezova L, Jiang BH, Flynn DC. The intrinsic ability of AFAP-110 to alter actin filament integrity is linked with its ability to also activate cellular tyrosine kinases. Oncogene. 2001a;20:6607–6616. - PubMed
-
- Baisden JM, Qian Y, Zot HM, Flynn DC. The actin filament-associated protein AFAP-110 is an adaptor protein that modulates changes in actin filament integrity. Oncogene. 2001b;20:6435–6447. - PubMed
-
- Braak E, Arai K, Braak H. Cerebellar involvement in Pick’s disease: affliction of mossy fibers, monodendritic brush cells, and dentate projection neurons. Exp Neurol. 1999;159:153–163. - PubMed
-
- DiNitto JP, Lambright DG. Membrane and juxtamembrane targeting by PH and PTB domains. Biochim Biophys Acta. 2006;1761:850–867. - PubMed
-
- Dorfleutner A, Cho Y, Vincent D, Cunnick J, Lin H, Weed SA, Stehlik C, Flynn DC. Phosphorylation of AFAP-110 affects podosome lifespan in A7r5 cells. J Cell Sci. 2008;121:2394–2405. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

