vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize
- PMID: 21335375
- PMCID: PMC3077783
- DOI: 10.1105/tpc.110.075267
vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize
Abstract
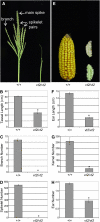
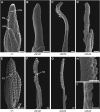
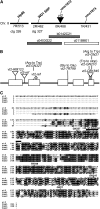
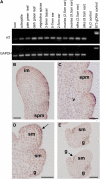
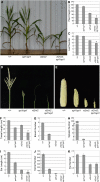
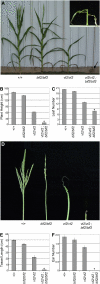
Auxin plays a fundamental role in organogenesis in plants. Multiple pathways for auxin biosynthesis have been proposed, but none of the predicted pathways are completely understood. Here, we report the positional cloning and characterization of the vanishing tassel2 (vt2) gene of maize (Zea mays). Phylogenetic analyses indicate that vt2 is a co-ortholog of TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1), which converts Trp to indole-3-pyruvic acid in one of four hypothesized Trp-dependent auxin biosynthesis pathways. Unlike single mutations in TAA1, which cause subtle morphological phenotypes in Arabidopsis thaliana, vt2 mutants have dramatic effects on vegetative and reproductive development. vt2 mutants share many similarities with sparse inflorescence1 (spi1) mutants in maize. spi1 is proposed to encode an enzyme in the tryptamine pathway for Trp-dependent auxin biosynthesis, although this biochemical activity has recently been questioned. Surprisingly, spi1 vt2 double mutants had only a slightly more severe phenotype than vt2 single mutants. Furthermore, both spi1 and vt2 single mutants exhibited a reduction in free auxin levels, but the spi1 vt2 double mutants did not have a further reduction compared with vt2 single mutants. Therefore, both spi1 and vt2 function in auxin biosynthesis in maize, possibly in the same pathway rather than independently as previously proposed.
Figures




References
-
- Barkawi L.S., Tam Y.Y., Tillman J.A., Normanly J., Cohen J.D. (2010). A high-throughput method for the quantitative analysis of auxins. Nat. Protoc. 5: 1609–1618 - PubMed
-
- Bartel B. (1997). Auxin biosynthesis. Annu. Rev. Plant Physiol. 48: 49–64 - PubMed
-
- Bartling D., Seedorf M., Mithöfer A., Weiler E.W. (1992). Cloning and expression of an Arabidopsis nitrilase which can convert indole-3-acetonitrile to the plant hormone, indole-3-acetic acid. Eur. J. Biochem. 205: 417–424 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

