Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling
- PMID: 21335377
- PMCID: PMC3077778
- DOI: 10.1105/tpc.110.082081
Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling
Abstract
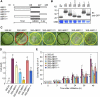
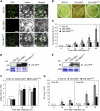
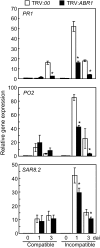
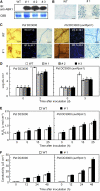
Abscisic acid (ABA) is a key regulator of plant growth and development, as well as plant defense responses. A high-throughput in planta proteome screen identified the pepper (Capsicum annuum) GRAM (for glucosyltransferases, Rab-like GTPase activators, and myotubularins) domain-containing ABA-RESPONSIVE1 (ABR1), which is highly induced by infection with avirulent Xanthomonas campestris pv vesicatoria and also by treatment with ABA. The GRAM domain is essential for the cell death response and for the nuclear localization of ABR1. ABR1 is required for priming cell death and reactive oxygen species production, as well as ABA-salicylic acid (SA) antagonism. Silencing of ABR1 significantly compromised the hypersensitive response but enhanced bacterial pathogen growth and ABA levels in pepper. High levels of ABA in ABR1-silenced plants antagonized the SA levels induced by pathogen infection. Heterologous transgenic expression of ABR1 in Arabidopsis thaliana conferred enhanced resistance to Pseudomonas syringae pv tomato and Hyaloperonospora arabidopsidis infection. The susceptibility of the Arabidopsis ABR1 putative ortholog mutant, abr1, to these pathogens also supports the involvement of ABR1 in disease resistance. Together, these results reveal ABR1 as a novel negative regulator of ABA signaling and suggest that the nuclear ABR1 pool is essential for the cell death induction associated with ABA-SA antagonism.
Figures








References
-
- Anderson J.P., Badruzsaufari E., Schenk P.M., Manners J.M., Desmond O.J., Ehlert C., Maclean D.J., Ebert P.R., Kazan K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 - PMC - PubMed
-
- Bahk Y.Y., Kim S.A., Kim J.S., Euh H.J., Bai G.H., Cho S.N., Kim Y.S. (2004). Antigens secreted from Mycobacterium tuberculosis: Identification by proteomics approach and test for diagnostic marker. Proteomics 4: 3299–3307 - PubMed
-
- Choi H.W., Kim Y.J., Hwang B.K. (2011). The hypersensitive induced reaction and leucine-rich repeat proteins regulate plant cell death associated with disease and plant immunity. Mol. Plant Microbe Interact. 24: 68–78 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

