A comparative analysis of the NADPH thioredoxin reductase C-2-Cys peroxiredoxin system from plants and cyanobacteria
- PMID: 21335525
- PMCID: PMC3091103
- DOI: 10.1104/pp.110.171082
A comparative analysis of the NADPH thioredoxin reductase C-2-Cys peroxiredoxin system from plants and cyanobacteria
Abstract
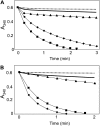
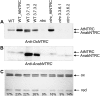
Redox regulation based on disulfide-dithiol conversion catalyzed by thioredoxins is an important component of chloroplast function. The reducing power is provided by ferredoxin reduced by the photosynthetic electron transport chain. In addition, chloroplasts are equipped with a peculiar NADPH-dependent thioredoxin reductase, termed NTRC, with a joint thioredoxin domain at the carboxyl terminus. Because NADPH can be produced by the oxidative pentose phosphate pathway during the night, NTRC is important to maintain the chloroplast redox homeostasis under light limitation. NTRC is exclusive for photosynthetic organisms such as plants, algae, and some, but not all, cyanobacteria. Phylogenetic analysis suggests that chloroplast NTRC originated from an ancestral cyanobacterial enzyme. While the biochemical properties of plant NTRC are well documented, little is known about the cyanobacterial enzyme. With the aim of comparing cyanobacterial and plant NTRCs, we have expressed the full-length enzyme from the cyanobacterium Anabaena species PCC 7120 as well as site-directed mutant variants and truncated polypeptides containing the NTR or the thioredoxin domains of the protein. Immunological and kinetic analysis showed a high similarity between NTRCs from plants and cyanobacteria. Both enzymes efficiently reduced 2-Cys peroxiredoxins from plants and from Anabaena but not from the cyanobacterium Synechocystis. Arabidopsis (Arabidopsis thaliana) NTRC knockout plants were transformed with the Anabaena NTRC gene. Despite a lower content of NTRC than in wild-type plants, the transgenic plants showed significant recovery of growth and pigmentation. Therefore, the Anabaena enzyme fulfills functions of the plant enzyme in vivo, further emphasizing the similarity between cyanobacterial and plant NTRCs.
Figures






Similar articles
-
Functional Significance of NADPH-Thioredoxin Reductase C in the Antioxidant Defense System of Cyanobacterium Anabaena sp. PCC 7120.Plant Cell Physiol. 2017 Jan 1;58(1):86-94. doi: 10.1093/pcp/pcw182. Plant Cell Physiol. 2017. PMID: 28011872
-
Molecular recognition in the interaction of chloroplast 2-Cys peroxiredoxin with NADPH-thioredoxin reductase C (NTRC) and thioredoxin x.FEBS Lett. 2014 Nov 28;588(23):4342-7. doi: 10.1016/j.febslet.2014.09.044. Epub 2014 Oct 16. FEBS Lett. 2014. PMID: 25448674
-
Chloroplast redox homeostasis is essential for lateral root formation in Arabidopsis.Plant Signal Behav. 2012 Sep 1;7(9):1177-9. doi: 10.4161/psb.21001. Epub 2012 Aug 17. Plant Signal Behav. 2012. PMID: 22899086 Free PMC article.
-
The function of the NADPH thioredoxin reductase C-2-Cys peroxiredoxin system in plastid redox regulation and signalling.FEBS Lett. 2012 Aug 31;586(18):2974-80. doi: 10.1016/j.febslet.2012.07.003. Epub 2012 Jul 10. FEBS Lett. 2012. PMID: 22796111 Review.
-
Thioredoxins and thioredoxin reductase in chloroplasts: A review.Gene. 2019 Jul 20;706:32-42. doi: 10.1016/j.gene.2019.04.041. Epub 2019 Apr 24. Gene. 2019. PMID: 31028868 Review.
Cited by
-
NADPH-Thioredoxin Reductase C Mediates the Response to Oxidative Stress and Thermotolerance in the Cyanobacterium Anabaena sp. PCC7120.Front Microbiol. 2016 Aug 18;7:1283. doi: 10.3389/fmicb.2016.01283. eCollection 2016. Front Microbiol. 2016. PMID: 27588019 Free PMC article.
-
Characterization of TrxC, an Atypical Thioredoxin Exclusively Present in Cyanobacteria.Antioxidants (Basel). 2018 Nov 13;7(11):164. doi: 10.3390/antiox7110164. Antioxidants (Basel). 2018. PMID: 30428557 Free PMC article.
-
Back to the future: Transplanting the chloroplast TrxF-FBPase-SBPase redox system to cyanobacteria.Front Plant Sci. 2022 Nov 28;13:1052019. doi: 10.3389/fpls.2022.1052019. eCollection 2022. Front Plant Sci. 2022. PMID: 36518499 Free PMC article.
-
Calredoxin regulates the chloroplast NADPH-dependent thioredoxin reductase in Chlamydomonas reinhardtii.Plant Physiol. 2023 Oct 26;193(3):2122-2140. doi: 10.1093/plphys/kiad426. Plant Physiol. 2023. PMID: 37474113 Free PMC article.
-
Exploring the Diversity of the Thioredoxin Systems in Cyanobacteria.Antioxidants (Basel). 2022 Mar 28;11(4):654. doi: 10.3390/antiox11040654. Antioxidants (Basel). 2022. PMID: 35453339 Free PMC article. Review.
References
-
- Baier M, Dietz K-J. (1997) The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J 12: 179–190 - PubMed
-
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 - PubMed
-
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

