Evolution of mating within the Candida parapsilosis species group
- PMID: 21335529
- PMCID: PMC3127640
- DOI: 10.1128/EC.00276-10
Evolution of mating within the Candida parapsilosis species group
Abstract
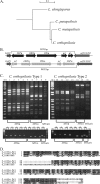
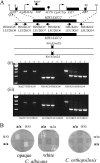
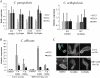
Candida orthopsilosis and Candida metapsilosis are closely related to Candida parapsilosis, a major cause of infection in premature neonates. Mating has not been observed in these species. We show that ∼190 isolates of C. parapsilosis contain only an MTLa idiomorph at the mating-type-like locus. Here, we describe the isolation and characterization of the MTL loci from C. orthopsilosis and C. metapsilosis. Among 16 C. orthopsilosis isolates, 9 were homozygous for MTLa, 5 were homozygous for MTLα, and 2 were MTLa/α heterozygotes. The C. orthopsilosis isolates belonged to two divergent groups, as characterized by restriction patterns at MTL, which probably represent subspecies. We sequenced both idiomorphs from each group and showed that they are 95% identical and that the regulatory genes are intact. In contrast, 18 isolates of C. metapsilosis contain only MTLα idiomorphs. Our results suggest that the role of MTL in determining cell type is being eroded in the C. parapsilosis species complex. The population structure of C. orthopsilosis indicates that mating may occur. However, expression of genes in the mating signal transduction pathway does not respond to exposure to alpha factor. C. parapsilosis is also nonresponsive, even when the GTPase-activating protein gene SST2 is deleted. In addition, splicing of introns in MTLa1 and MTLa2 is defective in C. orthopsilosis. Mating is not detected. The alpha factor peptide, which is the same sequence in C. parapsilosis, C. orthopsilosis, and C. metapsilosis, can induce a mating response in Candida albicans. It is therefore likely either that mating of C. orthopsilosis takes place under certain unidentified conditions or that the mating pathway has been adapted for other functions, such as cross-species communication.
Figures



Similar articles
-
A genome sequence survey shows that the pathogenic yeast Candida parapsilosis has a defective MTLa1 allele at its mating type locus.Eukaryot Cell. 2005 Jun;4(6):1009-17. doi: 10.1128/EC.4.6.1009-1017.2005. Eukaryot Cell. 2005. PMID: 15947193 Free PMC article.
-
MTL genotypes, phenotypic switching, and susceptibility profiles of Candida parapsilosis species group compared to Lodderomyces elongisporus.PLoS One. 2017 Aug 3;12(8):e0182653. doi: 10.1371/journal.pone.0182653. eCollection 2017. PLoS One. 2017. PMID: 28771588 Free PMC article.
-
Molecular epidemiology and antifungal susceptibility of Candida parapsilosis sensu stricto, Candida orthopsilosis, and Candida metapsilosis in Taiwan.Diagn Microbiol Infect Dis. 2010 Nov;68(3):284-92. doi: 10.1016/j.diagmicrobio.2010.07.004. Epub 2010 Sep 20. Diagn Microbiol Infect Dis. 2010. PMID: 20851551
-
Prospective multicenter study of the epidemiology, molecular identification, and antifungal susceptibility of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis isolated from patients with candidemia.Antimicrob Agents Chemother. 2011 Dec;55(12):5590-6. doi: 10.1128/AAC.00466-11. Epub 2011 Sep 19. Antimicrob Agents Chemother. 2011. PMID: 21930869 Free PMC article.
-
Through a glass opaquely: the biological significance of mating in Candida albicans.Curr Opin Microbiol. 2004 Dec;7(6):661-5. doi: 10.1016/j.mib.2004.10.003. Curr Opin Microbiol. 2004. PMID: 15556040 Review.
Cited by
-
Genome analysis of five recently described species of the CUG-Ser clade uncovers Candida theae as a new hybrid lineage with pathogenic potential in the Candida parapsilosis species complex.DNA Res. 2022 Feb 27;29(2):dsac010. doi: 10.1093/dnares/dsac010. DNA Res. 2022. PMID: 35438177 Free PMC article.
-
Molecular Investigation of the Fatal Bloodstream Candida orthopsilosis Infection Case following Gastrectomy.Int J Mol Sci. 2023 Mar 31;24(7):6541. doi: 10.3390/ijms24076541. Int J Mol Sci. 2023. PMID: 37047514 Free PMC article.
-
The APSES transcription factor Efg1 is a global regulator that controls morphogenesis and biofilm formation in Candida parapsilosis.Mol Microbiol. 2013 Oct;90(1):36-53. doi: 10.1111/mmi.12345. Epub 2013 Aug 15. Mol Microbiol. 2013. PMID: 23895281 Free PMC article.
-
Conserved and divergent roles of Bcr1 and CFEM proteins in Candida parapsilosis and Candida albicans.PLoS One. 2011;6(12):e28151. doi: 10.1371/journal.pone.0028151. Epub 2011 Dec 1. PLoS One. 2011. PMID: 22145027 Free PMC article.
-
Multiple Origins of the Pathogenic Yeast Candida orthopsilosis by Separate Hybridizations between Two Parental Species.PLoS Genet. 2016 Nov 2;12(11):e1006404. doi: 10.1371/journal.pgen.1006404. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27806045 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

