The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection
- PMID: 21335604
- PMCID: PMC3075711
- DOI: 10.1074/jbc.M110.213728
The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection
Abstract
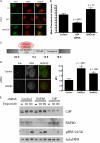
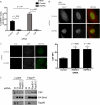
The tumor suppressor protein BRCA1 is a constituent of several different protein complexes and is required for homology-directed repair (HDR) of DNA double strand breaks (DSBs). The most recently discovered BRCA1-RAP80 complex is recruited to ubiquitin structures on chromatin surrounding the break. Deficiency of any member of this complex confers hypersensitivity to DNA-damaging agents by undefined mechanisms. In striking contrast to other BRCA1-containing complexes that are known to promote HDR, we demonstrate that the BRCA1-RAP80 complex restricts end resection in S/G(2) phase of the cell cycle, thereby limiting HDR. RAP80 or BRCC36 deficiency resulted in elevated Mre11-CtIP-dependent 5' end resection with a concomitant increase in HDR mechanisms that rely on 3' single-stranded overhangs. We propose a model in which the BRCA1-RAP80 complex limits nuclease accessibility to DSBs, thus preventing excessive end resection and potentially deleterious homology-directed DSB repair mechanisms that can impair genome integrity.
Figures




References
-
- Rahman N., Stratton M. R. (1998) Annu. Rev. Genet. 32, 95–121 - PubMed
-
- Scully R., Ganesan S., Vlasakova K., Chen J., Socolovsky M., Livingston D. M. (1999) Mol. Cell. 4, 1093–1099 - PubMed
-
- Stratton M. R., Rahman N. (2008) Nat. Genet. 40, 17–22 - PubMed
-
- Venkitaraman A. R. (2002) Cell 108, 171–182 - PubMed
-
- Karran P. (2000) Curr. Opin. Genet. Dev. 10, 144–150 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

