Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants
- PMID: 21335608
- PMCID: PMC3113581
- DOI: 10.1093/nar/gkr070
Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants
Abstract
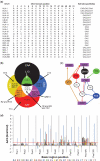
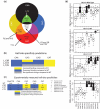
Numerous efforts are underway to determine gene regulatory networks that describe physical relationships between transcription factors (TFs) and their target DNA sequences. Members of paralogous TF families typically recognize similar DNA sequences. Knowledge of the molecular determinants of protein-DNA recognition by paralogous TFs is of central importance for understanding how small differences in DNA specificities can dictate target gene selection. Previously, we determined the in vitro DNA binding specificities of 19 Caenorhabditis elegans basic helix-loop-helix (bHLH) dimers using protein binding microarrays. These TFs bind E-box (CANNTG) and E-box-like sequences. Here, we combine these data with logics, bHLH-DNA co-crystal structures and computational modeling to infer which bHLH monomer can interact with which CAN E-box half-site and we identify a critical residue in the protein that dictates this specificity. Validation experiments using mutant bHLH proteins provide support for our inferences. Our study provides insights into the mechanisms of DNA recognition by bHLH dimers as well as a blueprint for system-level studies of the DNA binding determinants of other TF families in different model organisms and humans.
Figures


References
-
- Walhout AJM. Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome Res. 2006;16:1445–1454. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

