CESTA, a positive regulator of brassinosteroid biosynthesis
- PMID: 21336258
- PMCID: PMC3061039
- DOI: 10.1038/emboj.2011.35
CESTA, a positive regulator of brassinosteroid biosynthesis
Abstract
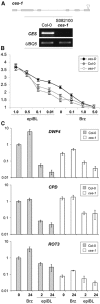
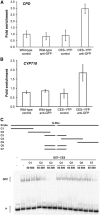
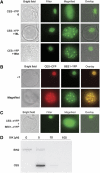
Brassinosteroids (BRs) are steroid hormones that are essential for the development of plants. A tight control of BR homeostasis is vital for modulating their impact on growth responses. Although it is recognized that the rapid adaptation of de novo synthesis has a key role in adjusting required BR levels, our knowledge of the mechanisms governing feedback control is limited. In this study, we identify the transcription factor CESTA as a regulator of BR biosynthesis. ces-D was isolated in a screen of Arabidopsis mutants by BR over-accumulation phenotypes. Loss-of-function analysis and the use of a dominant repressor version revealed functional overlap among CESTA and its homologues and confirmed the role of CESTA in the positive control of BR-biosynthetic gene expression. We provide evidence that CESTA interacts with its homologue BEE1 and can directly bind to a G-box motif in the promoter of the BR biosynthesis gene CPD. Moreover, we show that CESTA subnuclear localization is BR regulated and discuss a model, in which CESTA interplays with BEE1 to control BR biosynthesis and other BR responses.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Alonso JM, Stephanova AN, Leiss TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Aufsatz W (2005) Chromatin immunoprecipitation protocol to analyze histone modifications in Arabidopsis thaliana (PROT12) http://www.epigenome-noe.net/WWW/researchtools/protocol.php?protid=13
-
- Belkhadir Y, Chory J (2006) Brassinosteroid signaling: a paradigm for steroid hormone signaling from the cell surface. Science 314: 1410–1411 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

