Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics
- PMID: 21336272
- PMCID: PMC3501644
- DOI: 10.1038/nn.2765
Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics
Abstract
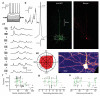
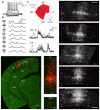
Single-cell genetic manipulation is expected to substantially advance the field of systems neuroscience. However, existing gene delivery techniques do not allow researchers to electrophysiologically characterize cells and to thereby establish an experimental link between physiology and genetics for understanding neuronal function. In the mouse brain in vivo, we found that neurons remained intact after 'blind' whole-cell recording, that DNA vectors could be delivered through the patch-pipette during such recordings and that these vectors drove protein expression in recorded cells for at least 7 d. To illustrate the utility of this approach, we recorded visually evoked synaptic responses in primary visual cortical cells while delivering DNA plasmids that allowed retrograde, monosynaptic tracing of each neuron's presynaptic inputs. By providing a biophysical profile of a cell before its specific genetic perturbation, this combinatorial method captures the synaptic and anatomical receptive field of a neuron.
Figures


References
-
- Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2:932–940. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

