In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-kappaB and activation of caspase-8
- PMID: 21339290
- PMCID: PMC3075675
- DOI: 10.1074/jbc.M110.216226
In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-kappaB and activation of caspase-8
Erratum in
-
In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-κB and activation of caspase-8.J Biol Chem. 2016 Jan 29;291(5):2547. doi: 10.1074/jbc.A110.216226. J Biol Chem. 2016. PMID: 26826208 Free PMC article. No abstract available.
Abstract
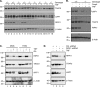
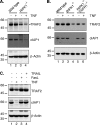
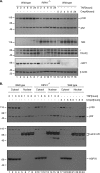
RIPK1 is involved in signaling from TNF and TLR family receptors. After receptor ligation, RIPK1 not only modulates activation of both canonical and NIK-dependent NF-κB, but also regulates caspase-8 activation and cell death. Although overexpression of RIPK1 can cause caspase-8-dependent cell death, when RIPK1(-/-) cells are exposed to TNF and low doses of cycloheximide, they die more readily than wild-type cells, indicating RIPK1 has pro-survival as well as pro-apoptotic activities. To determine how RIPK1 promotes cell survival, we compared wild-type and RIPK1(-/-) cells treated with TNF. Although TRAF2 levels remained constant in TNF-treated wild-type cells, TNF stimulation of RIPK1(-/-) cells caused TRAF2 and cIAP1 to be rapidly degraded by the proteasome, which led to an increase in NIK levels. This resulted in processing of p100 NF-κB2 to p52, a decrease in levels of cFLIP(L), and activation of caspase-8, culminating in cell death. Therefore, the pro-survival effect of RIPK1 is mediated by stabilization of TRAF2 and cIAP1.
Figures


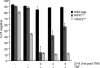


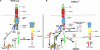
References
-
- Wong W. W., Gentle I. E., Nachbur U., Anderton H., Vaux D. L., Silke J. (2010) Cell Death Differ. 17, 482–487 - PubMed
-
- Kelliher M. A., Grimm S., Ishida Y., Kuo F., Stanger B. Z., Leder P. (1998) Immunity 8, 297–303 - PubMed
-
- Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Barker P. A. (2008) Mol. Cell 30, 689–700 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

