Inhibitory signaling blocks activating receptor clustering and induces cytoskeletal retraction in natural killer cells
- PMID: 21339333
- PMCID: PMC3044118
- DOI: 10.1083/jcb.201009135
Inhibitory signaling blocks activating receptor clustering and induces cytoskeletal retraction in natural killer cells
Abstract
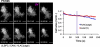
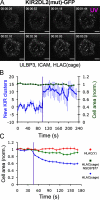
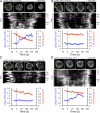
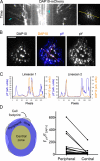
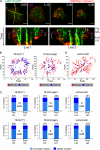
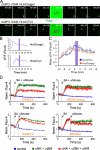
Natural killer (NK) lymphocytes use a variety of activating receptors to recognize and kill infected or tumorigenic cells during an innate immune response. To prevent targeting healthy tissue, NK cells also express numerous inhibitory receptors that signal through immunotyrosine-based inhibitory motifs (ITIMs). Precisely how signals from competing activating and inhibitory receptors are integrated and resolved is not understood. To investigate how ITIM receptor signaling impinges on activating pathways, we developed a photochemical approach for stimulating the inhibitory receptor KIR2DL2 during ongoing NK cell-activating responses in high-resolution imaging experiments. Photostimulation of KIR2DL2 induces the rapid formation of inhibitory receptor microclusters in the plasma membrane and the simultaneous suppression of microclusters containing activating receptors. This is followed by the collapse of the peripheral actin cytoskeleton and retraction of the NK cell from the source of inhibitory stimulation. These results suggest a cell biological basis for ITIM receptor signaling and establish an experimental framework for analyzing it.
Figures




References
-
- Barber D.F., Faure M., Long E.O. 2004. LFA-1 contributes an early signal for NK cell cytotoxicity. J. Immunol. 173:3653–3659 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

