SoftWAXS: a computational tool for modeling wide-angle X-ray solution scattering from biomolecules
- PMID: 21339902
- PMCID: PMC3041499
- DOI: 10.1107/S0021889809032919
SoftWAXS: a computational tool for modeling wide-angle X-ray solution scattering from biomolecules
Abstract
This paper describes a computational approach to estimating wide-angle X-ray solution scattering (WAXS) from proteins, which has been implemented in a computer program called SoftWAXS. The accuracy and efficiency of SoftWAXS are analyzed for analytically solvable model problems as well as for proteins. Key features of the approach include a numerical procedure for performing the required spherical averaging and explicit representation of the solute-solvent boundary and the surface of the hydration layer. These features allow the Fourier transform of the excluded volume and hydration layer to be computed directly and with high accuracy. This approach will allow future investigation of different treatments of the electron density in the hydration shell. Numerical results illustrate the differences between this approach to modeling the excluded volume and a widely used model that treats the excluded-volume function as a sum of Gaussians representing the individual atomic excluded volumes. Comparison of the results obtained here with those from explicit-solvent molecular dynamics clarifies shortcomings inherent to the representation of solvent as a time-averaged electron-density profile. In addition, an assessment is made of how the calculated scattering patterns depend on input parameters such as the solute-atom radii, the width of the hydration shell and the hydration-layer contrast. These results suggest that obtaining predictive calculations of high-resolution WAXS patterns may require sophisticated treatments of solvent.
Figures



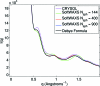
 is in Å
is in Å . Units on the ordinate axis are arbitrary intensity units.
. Units on the ordinate axis are arbitrary intensity units.
 Å
Å ) due to the use of coarser cubes. Units on the ordinate axis are arbitrary intensity units. Excluded-volume contributions to
) due to the use of coarser cubes. Units on the ordinate axis are arbitrary intensity units. Excluded-volume contributions to  are as follows: for 5-level octree,
are as follows: for 5-level octree,  ; for 6-level octree,
; for 6-level octree,  ; for CRYSOL,
; for CRYSOL,  .
.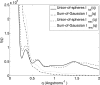
 . Using this approach the excluded-volume contribution is
. Using this approach the excluded-volume contribution is  . See §4.3.4 for more details. The sum-of-Gaussians calculation employed the same atomic excluded volumes as used in CRYSOL: C 16.44, O 9.13, N 2.49, H 5.15 Å
. See §4.3.4 for more details. The sum-of-Gaussians calculation employed the same atomic excluded volumes as used in CRYSOL: C 16.44, O 9.13, N 2.49, H 5.15 Å (Fraser et al., 1978; Svergun et al., 1995 ▶). The excluded-volume contribution is
(Fraser et al., 1978; Svergun et al., 1995 ▶). The excluded-volume contribution is  .
.
 Å
Å , where the semilog scale provides adequate resolution to observe the minute discrepancy.
, where the semilog scale provides adequate resolution to observe the minute discrepancy.
 and contrast
and contrast  for
for  Å
Å . For larger
. For larger  the parameters, when varied within reasonable limits, have relatively little effect on the pattern. In each plot the products of hydration-layer density (relative to bulk water) and thickness are the same for the two patterns. (a)
the parameters, when varied within reasonable limits, have relatively little effect on the pattern. In each plot the products of hydration-layer density (relative to bulk water) and thickness are the same for the two patterns. (a)  e Å−2, (b)
e Å−2, (b)  e Å−2, (c)
e Å−2, (c)  e Å−2, (d)
e Å−2, (d)  e Å−2.
e Å−2.

References
-
- Brooks, B. R., Bruccoleri, R. E., Olafson, B. D., States, D. J., Swaminathan, S. & Karplus, M. (1983). J. Comput. Chem. 4, 187–217.
Grants and funding
LinkOut - more resources
Full Text Sources
