Phytophthora taxa associated with cultivated Agathosma, with emphasis on the P. citricola complex and P. capensis sp. nov
- PMID: 21339965
- PMCID: PMC3028514
- DOI: 10.3767/003158510X538371
Phytophthora taxa associated with cultivated Agathosma, with emphasis on the P. citricola complex and P. capensis sp. nov
Abstract
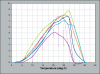
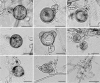
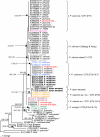
Agathosma species, which are indigenous to South Africa, are also cultivated for commercial use. Recently growers experienced severe plant loss, and symptoms shown by affected plants suggested that a soilborne disease could be the cause of death. A number of Phytophthora taxa were isolated from diseased plants, and this paper reports their identity, mating type, and pathogenicity to young Agathosma plants. Using morphological and sequence data seven Phytophthora taxa were identified: the A1 mating type of P. cinnamomi var. cinnamomi, P. cinnamomi var. parvispora and P. cryptogea, the A2 mating type of P. drechsleri and P. nicotianae, and two homothallic taxa from the P. citricola complex. The identity of isolates in the P. citricola complex was resolved using reference isolates of P. citricola CIT groups 1 to 5 sensu Oudemans et al. (1994) along with multi-locus phylogenies (three nuclear and two mitochondrial regions), isozyme analyses, morphological characteristics and temperature-growth studies. These analyses revealed the isolates from Agathosma to include P. multivora and a putative novel species, P. taxon emzansi. Furthermore, among the P. citricola reference isolates the presence of a new species was revealed, described here as P. capensis. Findings of our study, along with some recent other studies, have contributed to resolving some of the species complexity within the P. citricola complex, resulting in the identification of a number of phylogenetically distinct taxa. The pathogenicity of representative isolates of the taxa from Agathosma was tested on A. betulina seedlings. The putative novel species, P. taxon emzansi, and P. cinnamomi var. parvispora were non-pathogenic, whereas the other species were pathogenic to this host.
Keywords: avocado; buchu; fynbos; glucose-6-phosphate isomerase; isozymes; malate dehydrogenase; pathogenicity; root-rot; taxonomy.
Figures



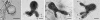


References
-
- Abad ZG, Abad JA, Coffey MD, Oudemans PV, Man in ‘tVeld WA , et al. 2008. Phytophthora bisheria sp. nov., a new species identified in isolates from the Rosaceous raspberry, rose and strawberry in three continents. Mycologia 100: 99 – 110 - PubMed
-
- Álvarez I, Wendel JF.2003. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution 29: 417 – 434 - PubMed
-
- Bailey CD, Carr TG, Harris SA, Hughes CE.2003. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Molecular Phylogenetics and Evolution 29: 435 – 455 - PubMed
-
- Blair JE, Coffey MD, Park S-Y, Geiser DM, Kang S.2008. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genetics and Biology 45: 266 – 277 - PubMed
-
- Boersma JG, Cooke DEL, Sivasithamparam K.2000. A survey of wildflower farms in the south-west of Western Australia for Phytophthora spp. associated with root rots. Australian Journal of Experimental Agriculture 40: 1011 – 1019
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
