Detection of Mycosphaerella graminicola in wheat leaves by a microsatellite dinucleotide specific-primer
- PMID: 21340008
- PMCID: PMC3039974
- DOI: 10.3390/ijms12010682
Detection of Mycosphaerella graminicola in wheat leaves by a microsatellite dinucleotide specific-primer
Abstract
Early detection of infection is very important for efficient management of Mycosphaerella graminicola leaf blotch. To monitor and quantify the occurrence of this fungus during the growing season, a diagnostic method based on real-time PCR was developed. Standard and real-time PCR assays were developed using SYBR Green chemistry to quantify M. graminicola in vitro or in wheat samples. Microsatellite dinucleotide specific-primers were designed based on microsatellite repeats of sequences present in the genome of M. graminicola. Specificity was checked by analyzing DNA of 55 M. graminicola isolates obtained from different geographical origins. The method appears to be highly specific for detecting M. graminicola; no fluorescent signals were observed from 14 other closely related taxa. Primer (CT) 7 G amplified a specific amplicon of 570 bp from all M. graminicola isolates. The primers did not amplify DNA extracted from 14 other fungal species. The approximate melting temperature (Tm) of the (CT) 7 G primer was 84.2 °C. The detection limit of the real-time PCR assay with the primer sets (CT) 7 G is 10 fg/25 μL, as compared to 10 pg/25 μL using conventional PCR technology. From symptomless leaves, a PCR fragment could be generated two days after inoculation. Both conventional and real-time PCR could successfully detect the fungus from artificially inoculated wheat leaves. However, real-time PCR appeared much more sensitive than conventional PCR. The developed quantitative real-time PCR method proved to be rapid, sensitive, specific, cost-effective and reliable for the identification and quantification of M. graminicola in wheat.
Keywords: Dothidiomycete; Septoria tritici blotch; microsatellite; molecular diagnostics; wheat.
Figures


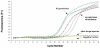

Similar articles
-
Identification and genetic mapping of highly polymorphic microsatellite loci from an EST database of the septoria tritici blotch pathogen Mycosphaerella graminicola.Fungal Genet Biol. 2007 May;44(5):398-414. doi: 10.1016/j.fgb.2006.09.004. Epub 2006 Oct 30. Fungal Genet Biol. 2007. PMID: 17074520
-
Presymptomatic and quantitative detection of Mycosphaerella graminicola development in wheat using a real-time PCR assay.FEMS Microbiol Lett. 2006 Sep;262(2):223-9. doi: 10.1111/j.1574-6968.2006.00393.x. FEMS Microbiol Lett. 2006. PMID: 16923079
-
Genetic differentiation at microsatellite loci among populations of Mycosphaerella graminicola from California, Indiana, Kansas, and North Dakota.Phytopathology. 2011 Oct;101(10):1251-9. doi: 10.1094/PHYTO-08-10-0212. Phytopathology. 2011. PMID: 21692645
-
Chromosomal location of a race-specific resistance gene to Mycosphaerella graminicola in the spring wheat ST6.Theor Appl Genet. 2003 Nov;107(7):1181-6. doi: 10.1007/s00122-003-1359-0. Epub 2003 Jul 26. Theor Appl Genet. 2003. PMID: 12898022
-
Cell biology of Zymoseptoria tritici: Pathogen cell organization and wheat infection.Fungal Genet Biol. 2015 Jun;79:17-23. doi: 10.1016/j.fgb.2015.04.002. Fungal Genet Biol. 2015. PMID: 26092785 Free PMC article. Review.
Cited by
-
Species-Specific Detection of Mycosphaerella polygoni-cuspidati as a Biological Control Agent for Fallopia japonica by PCR Assay.Mol Biotechnol. 2016 Oct;58(10):626-633. doi: 10.1007/s12033-016-9962-x. Mol Biotechnol. 2016. PMID: 27389682 Free PMC article.
-
Promising Perspectives for Detection, Identification, and Quantification of Plant Pathogenic Fungi and Oomycetes through Targeting Mitochondrial DNA.Int J Mol Sci. 2020 Apr 10;21(7):2645. doi: 10.3390/ijms21072645. Int J Mol Sci. 2020. PMID: 32290169 Free PMC article. Review.
-
Evaluation of FLASH - PCR forrapid detection of Mycobacterium tuberculosis from clinical specimens.Iran J Microbiol. 2013 Dec;5(4):383-90. Iran J Microbiol. 2013. PMID: 25848509 Free PMC article.
References
-
- Eyal Z. The Septoria tritici and Stagonospora nodorum blotch diseases of wheat. Eur. J. Plant Pathol. 1999;105:629–641.
-
- Desmazières JBHJ. Cryptogames nouvelles. Ann. Sci. Nat. 1842;17:91–118.
-
- Sanderson FR. A Mycosphaerella species as the ascogenous state of Septoria tritici Rob. and Desm. NZ J. Bot. 1972;10:707–709.
-
- Sanderson FR. Mycosphaerella graminicola (Fuckel) Sand-erson comb. nov., the ascogenous state of Septoria tritici Rob. and Desm. NZ J. Bot. 1976;14:359–360.
-
- Halama P. The occurrence of Mycosphaerella graminicola, teleomorph of Septoria tritici in France. Plant Pathol. 1996;45:135–138.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

