Constrained proper sampling of conformations of transition state ensemble of protein folding
- PMID: 21341875
- PMCID: PMC3071304
- DOI: 10.1063/1.3519056
Constrained proper sampling of conformations of transition state ensemble of protein folding
Abstract
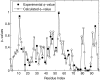
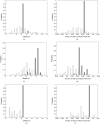
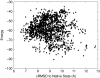
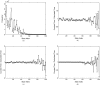
Characterizing the conformations of protein in the transition state ensemble (TSE) is important for studying protein folding. A promising approach pioneered by Vendruscolo et al. [Nature (London) 409, 641 (2001)] to study TSE is to generate conformations that satisfy all constraints imposed by the experimentally measured φ values that provide information about the native likeness of the transition states. Faísca et al. [J. Chem. Phys. 129, 095108 (2008)] generated conformations of TSE based on the criterion that, starting from a TS conformation, the probabilities of folding and unfolding are about equal through Markov Chain Monte Carlo (MCMC) simulations. In this study, we use the technique of constrained sequential Monte Carlo method [Lin et al., J. Chem. Phys. 129, 094101 (2008); Zhang et al. Proteins 66, 61 (2007)] to generate TSE conformations of acylphosphatase of 98 residues that satisfy the φ-value constraints, as well as the criterion that each conformation has a folding probability of 0.5 by Monte Carlo simulations. We adopt a two stage process and first generate 5000 contact maps satisfying the φ-value constraints. Each contact map is then used to generate 1000 properly weighted conformations. After clustering similar conformations, we obtain a set of properly weighted samples of 4185 candidate clusters. Representative conformation of each of these cluster is then selected and 50 runs of Markov chain Monte Carlo (MCMC) simulation are carried using a regrowth move set. We then select a subset of 1501 conformations that have equal probabilities to fold and to unfold as the set of TSE. These 1501 samples characterize well the distribution of transition state ensemble conformations of acylphosphatase. Compared with previous studies, our approach can access much wider conformational space and can objectively generate conformations that satisfy the φ-value constraints and the criterion of 0.5 folding probability without bias. In contrast to previous studies, our results show that transition state conformations are very diverse and are far from nativelike when measured in cartesian root-mean-square deviation (cRMSD): the average cRMSD between TSE conformations and the native structure is 9.4 Å for this short protein, instead of 6 Å reported in previous studies. In addition, we found that the average fraction of native contacts in the TSE is 0.37, with enrichment in native-like β-sheets and a shortage of long range contacts, suggesting such contacts form at a later stage of folding. We further calculate the first passage time of folding of TSE conformations through calculation of physical time associated with the regrowth moves in MCMC simulation through mapping such moves to a Markovian state model, whose transition time was obtained by Langevin dynamics simulations. Our results indicate that despite the large structural diversity of the TSE, they are characterized by similar folding time. Our approach is general and can be used to study TSE in other macromolecules.
Figures

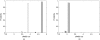
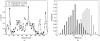


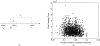

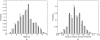
Similar articles
-
Three key residues form a critical contact network in a protein folding transition state.Nature. 2001 Feb 1;409(6820):641-5. doi: 10.1038/35054591. Nature. 2001. PMID: 11214326
-
Factors governing the foldability of proteins.Proteins. 1996 Dec;26(4):411-41. doi: 10.1002/(SICI)1097-0134(199612)26:4<411::AID-PROT4>3.0.CO;2-E. Proteins. 1996. PMID: 8990496
-
Reconstruction of the src-SH3 protein domain transition state ensemble using multiscale molecular dynamics simulations.J Mol Biol. 2005 Jul 29;350(5):1035-50. doi: 10.1016/j.jmb.2005.05.017. J Mol Biol. 2005. PMID: 15982666
-
Dynamic Monte Carlo simulations of a new lattice model of globular protein folding, structure and dynamics.J Mol Biol. 1991 Sep 20;221(2):499-531. doi: 10.1016/0022-2836(91)80070-b. J Mol Biol. 1991. PMID: 1920430 Review.
-
From covalent transition states in chemistry to noncovalent in biology: from β- to Φ-value analysis of protein folding.Q Rev Biophys. 2024 Mar 20;57:e4. doi: 10.1017/S0033583523000045. Q Rev Biophys. 2024. PMID: 38597675 Review.
Cited by
-
Conformational sampling and structure prediction of multiple interacting loops in soluble and β-barrel membrane proteins using multi-loop distance-guided chain-growth Monte Carlo method.Bioinformatics. 2015 Aug 15;31(16):2646-52. doi: 10.1093/bioinformatics/btv198. Epub 2015 Apr 9. Bioinformatics. 2015. PMID: 25861965 Free PMC article.
-
Computational studies of membrane proteins: models and predictions for biological understanding.Biochim Biophys Acta. 2012 Apr;1818(4):927-41. doi: 10.1016/j.bbamem.2011.09.026. Epub 2011 Oct 12. Biochim Biophys Acta. 2012. PMID: 22051023 Free PMC article. Review.
-
Spatial confinement is a major determinant of the folding landscape of human chromosomes.Nucleic Acids Res. 2014 Jul;42(13):8223-30. doi: 10.1093/nar/gku462. Epub 2014 Jul 2. Nucleic Acids Res. 2014. PMID: 24990374 Free PMC article.
-
Multiscale Modeling of Cellular Epigenetic States: Stochasticity in Molecular Networks, Chromatin Folding in Cell Nuclei, and Tissue Pattern Formation of Cells.Crit Rev Biomed Eng. 2015;43(4):323-46. doi: 10.1615/CritRevBiomedEng.2016016559. Crit Rev Biomed Eng. 2015. PMID: 27480462 Free PMC article.
-
Structure Prediction of RNA Loops with a Probabilistic Approach.PLoS Comput Biol. 2016 Aug 5;12(8):e1005032. doi: 10.1371/journal.pcbi.1005032. eCollection 2016 Aug. PLoS Comput Biol. 2016. PMID: 27494763 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

