L3MBTL1 deficiency directs the differentiation of human embryonic stem cells toward trophectoderm
- PMID: 21341991
- PMCID: PMC3202892
- DOI: 10.1089/scd.2010.0437
L3MBTL1 deficiency directs the differentiation of human embryonic stem cells toward trophectoderm
Abstract
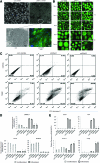
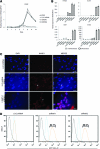
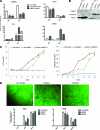
Human embryonic stem cells (hESCs) can be used to study the early events in human development and, hopefully, to understand how to differentiate human pluripotent cells for clinical use. To define how L3MBTL1, a chromatin-associated polycomb group protein with transcriptional repressive activities, regulates early events in embryonic cell differentiation, we created hESC lines that constitutively express shRNAs directed against L3MBTL1. The L3MBTL1 knockdown (KD) hESCs maintained normal morphology, proliferation, cell cycle kinetics, cell surface markers, and karyotype after 40 passages. However, under conditions that promote spontaneous differentiation, the L3MBTL1 KD cells differentiated into a relatively homogeneous population of large, flat trophoblast-like cells, unlike the multilineage differentiation seen with the control cells. The differentiated L3MBTL1 KD cells expressed numerous trophoblast markers and secreted placental hormones. Although the L3MBTL1 KD cells could be induced to differentiate into various embryonic lineages, they adopted an exclusive trophoblast fate during spontaneous differentiation. Our data demonstrate that depletion of L3MBTL1 does not affect hESC self-renewal, rather it enhances differentiation toward extra-embryonic trophoblast tissues.
Figures

References
-
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. - PubMed
-
- Auman HJ. Nottoli T. Lakiza O. Winger Q. Donaldson S. Williams T. Transcription factor AP-2gamma is essential in the extra-embryonic lineages for early postimplantation development. Development. 2002;129:2733–2747. - PubMed
-
- Donnison M. Beaton A. Davey HW. Broadhurst R. L'Huillier P. Pfeffer PL. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development. 2005;132:2299–2308. - PubMed
-
- Ralston A. Cox BJ. Nishioka N. Sasaki H. Chea E. Rugg-Gunn P. Guo G. Robson P. Draper JS. Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 137:395–403. - PubMed
-
- Russ AP. Wattler S. Colledge WH. Aparicio SA. Carlton MB. Pearce JJ. Barton SC. Surani MA. Ryan K. Nehls MC. Wilson V. Evans MJ. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

