pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes
- PMID: 21342504
- PMCID: PMC3056738
- DOI: 10.1186/1471-2180-11-38
pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes
Abstract
Background: Since publication in 1977 of plasmid pBR322, many breakthroughs in Biology have depended on increasingly sophisticated vector platforms for analysis and engineering of given bacterial strains. Although restriction sites impose a certain format in the procedures for assembling cloned genes, every attempt thus far to standardize vector architecture and nomenclature has ended up in failure. While this state of affairs may still be tolerable for traditional one-at-a-time studies of single genes, the onset of systems and synthetic biology calls for a simplification--along with an optimization--of the currently unwieldy pool of genetic tools.
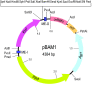
Results: The functional DNA sequences present in the natural bacterial transposon Tn5 have been methodically edited and refactored for the production of a multi-purpose genetic tool named pBAM1, which allows a range of manipulations in the genome of gram-negative bacteria. This all-synthetic construct enhances the power of mini-transposon vectors for either de-construction or re-construction of phenotypes á la carte by incorporating features inspired in systems engineering: modularity, re-usability, minimization, and compatibility with other genetic tools. pBAM1 bears an streamlined, restriction site-freed and narrow-host range replication frame bearing the sequences of R6K oriV, oriT and an ampicillin resistance marker. These go along with a business module that contains a host-independent and hyperactive transposition platform for in vivo or in vitro insertion of desired DNA into the genome of the target bacterium. All functional sequences were standardized for a straightforward replacement by equivalent counterparts, if required. pBAM1 can be delivered into recipient cells by either mating or electroporation, producing transposon insertion frequencies of 1.8 × 10(-3) and 1.02 × 10(-7), respectively in the soil bacterium Pseudomonas putida. Analyses of the resulting clones revealed a 100% of unique transposition events and virtually no-cointegration of the donor plasmid within the target genome.
Conclusions: This work reports the design and performance of an all-synthetic mini-transposon vector. The power of the new system for both identification of new functions or for the construction of desired phenotypes is shown in a genetic survey of hyper-expressed proteins and regulatory elements that influence the expression of the σ54-dependent Pu promoter of P. putida.
Figures

Similar articles
-
Engineering Gram-Negative Microbial Cell Factories Using Transposon Vectors.Methods Mol Biol. 2017;1498:273-293. doi: 10.1007/978-1-4939-6472-7_18. Methods Mol Biol. 2017. PMID: 27709582
-
Transposon-based and plasmid-based genetic tools for editing genomes of gram-negative bacteria.Methods Mol Biol. 2012;813:267-83. doi: 10.1007/978-1-61779-412-4_16. Methods Mol Biol. 2012. PMID: 22083748
-
Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria.J Bacteriol. 1990 Nov;172(11):6557-67. doi: 10.1128/jb.172.11.6557-6567.1990. J Bacteriol. 1990. PMID: 2172216 Free PMC article.
-
Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans-metabolism.Metab Eng. 2018 Nov;50:142-155. doi: 10.1016/j.ymben.2018.05.005. Epub 2018 May 16. Metab Eng. 2018. PMID: 29758287 Review.
-
A navigation guide of synthetic biology tools for Pseudomonas putida.Biotechnol Adv. 2021 Jul-Aug;49:107732. doi: 10.1016/j.biotechadv.2021.107732. Epub 2021 Mar 27. Biotechnol Adv. 2021. PMID: 33785373 Review.
Cited by
-
Refactoring the Conjugation Machinery of Promiscuous Plasmid RP4 into a Device for Conversion of Gram-Negative Isolates to Hfr Strains.ACS Synth Biol. 2021 Apr 16;10(4):690-697. doi: 10.1021/acssynbio.0c00611. Epub 2021 Mar 22. ACS Synth Biol. 2021. PMID: 33750103 Free PMC article.
-
Stepwise genetic engineering of Pseudomonas putida enables robust heterologous production of prodigiosin and glidobactin A.Metab Eng. 2021 Sep;67:112-124. doi: 10.1016/j.ymben.2021.06.004. Epub 2021 Jun 24. Metab Eng. 2021. PMID: 34175462 Free PMC article.
-
Random and cyclical deletion of large DNA segments in the genome of Pseudomonas putida.Environ Microbiol. 2012 Jun;14(6):1444-53. doi: 10.1111/j.1462-2920.2012.02730.x. Epub 2012 Mar 19. Environ Microbiol. 2012. PMID: 22429517 Free PMC article.
-
Production of selenium nanoparticles occurs through an interconnected pathway of sulphur metabolism and oxidative stress response in Pseudomonas putida KT2440.Microb Biotechnol. 2023 May;16(5):931-946. doi: 10.1111/1751-7915.14215. Epub 2023 Jan 22. Microb Biotechnol. 2023. PMID: 36682039 Free PMC article.
-
Bimodal Expression of the Salmonella Typhimurium spv Operon.Genetics. 2018 Oct;210(2):621-635. doi: 10.1534/genetics.118.300822. Epub 2018 Aug 16. Genetics. 2018. PMID: 30143595 Free PMC article.
References
-
- de Lorenzo V, Herrero M, Sánchez JM, Timmis KN. Mini-transposons in microbial ecology and environmental biotechnology. FEMS Microbiology Ecology. 1998;27:211–224. doi: 10.1016/S0168-6496(98)00064-6. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

