Peptide inhibition of human cytomegalovirus infection
- PMID: 21342525
- PMCID: PMC3050824
- DOI: 10.1186/1743-422X-8-76
Peptide inhibition of human cytomegalovirus infection
Abstract
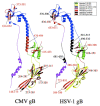
Background: Human cytomegalovirus (HCMV) is the most prevalent congenital viral infection in the United States and Europe causing significant morbidity and mortality to both mother and child. HCMV is also an opportunistic pathogen in immunocompromised individuals, including human immunodeficiency virus (HIV)- infected patients with AIDS, and solid organ and allogeneic stem cell transplantation recipients. Current treatments for HCMV-associated diseases are insufficient due to the emergence of drug-induced resistance and cytotoxicity, necessitating novel approaches to limit HCMV infection. The aim of this study was to develop therapeutic peptides targeting glycoprotein B (gB), a major glycoprotein of HCMV that is highly conserved across the Herpesviridae family, that specifically inhibit fusion of the viral envelope with the host cell membrane preventing HCMV entry and infection.
Results: Using the Wimley-White Interfacial Hydrophobicity Scale (WWIHS), several regions within gB were identified that display a high potential to interact with lipid bilayers of cell membranes and hydrophobic surfaces within proteins. The ability of synthetic peptides analogous to WWIHS-positive sequences of HCMV gB to inhibit viral infectivity was evaluated. Human foreskin fibroblasts (HFF) were infected with the Towne-GFP strain of HCMV (0.5 MOI), preincubated with peptides at a range of concentrations (78 nm to 100 μM), and GFP-positive cells were visualized 48 hours post-infection by fluorescence microscopy and analyzed quantitatively by flow cytometry. Peptides that inhibited HCMV infection demonstrated different inhibitory concentration curves indicating that each peptide possesses distinct biophysical properties. Peptide 174-200 showed 80% inhibition of viral infection at a concentration of 100 μM, and 51% and 62% inhibition at concentrations of 5 μM and 2.5 μM, respectively. Peptide 233-263 inhibited infection by 97% and 92% at concentrations of 100 μM and 50 μM, respectively, and 60% at a concentration of 2.5 μM. While peptides 264-291 and 297-315, individually failed to inhibit viral infection, when combined, they showed 67% inhibition of HCMV infection at a concentration of 0.125 μM each.
Conclusions: Peptides designed to target putative fusogenic domains of gB provide a basis for the development of novel therapeutics that prevent HCMV infection.
Figures


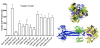
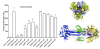
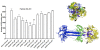
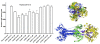
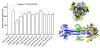
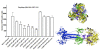

References
-
- Bate SL, Cannon MJ. A social marketing approach to building a behavioural intervention for congenital cytomegalovirus. Health Promot Pract. 2009. in press . - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

