Changed epitopes drive the antigenic drift for influenza A (H3N2) viruses
- PMID: 21342562
- PMCID: PMC3044287
- DOI: 10.1186/1471-2105-12-S1-S31
Changed epitopes drive the antigenic drift for influenza A (H3N2) viruses
Abstract
Background: In circulating influenza viruses, gradually accumulated mutations on the glycoprotein hemagglutinin (HA), which interacts with infectivity-neutralizing antibodies, lead to the escape of immune system (called antigenic drift). The antibody recognition is highly correlated to the conformation change on the antigenic sites (epitopes), which locate on HA surface. To quantify a changed epitope for escaping from neutralizing antibodies is the basis for the antigenic drift and vaccine development.
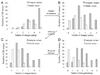
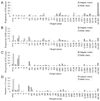
Results: We have developed an epitope-based method to identify the antigenic drift of influenza A utilizing the conformation changes on epitopes. A changed epitope, an antigenic site on HA with an accumulated conformation change to escape from neutralizing antibody, can be considered as a "key feature" for representing the antigenic drift. According to hemagglutination inhibition (HI) assays and HA/antibody complex structures, we statistically measured the conformation change of an epitope by considering the number of critical position mutations with high genetic diversity and antigenic scores. Experimental results show that two critical position mutations can induce the conformation change of an epitope to escape from the antibody recognition. Among five epitopes of HA, epitopes A and B, which are near to the receptor binding site, play a key role for neutralizing antibodies. In addition, two changed epitopes often drive the antigenic drift and can explain the selections of 24 WHO vaccine strains.
Conclusions: Our method is able to quantify the changed epitopes on HA for predicting the antigenic variants and providing biological insights to the vaccine updates. We believe that our method is robust and useful for studying influenza virus evolution and vaccine development.
Figures

Similar articles
-
Antigenic sites of H1N1 influenza virus hemagglutinin revealed by natural isolates and inhibition assays.Vaccine. 2012 Sep 28;30(44):6327-37. doi: 10.1016/j.vaccine.2012.07.079. Epub 2012 Aug 8. Vaccine. 2012. PMID: 22885274
-
Mapping of a Novel H3-Specific Broadly Neutralizing Monoclonal Antibody Targeting the Hemagglutinin Globular Head Isolated from an Elite Influenza Virus-Immunized Donor Exhibiting Serological Breadth.J Virol. 2020 Feb 28;94(6):e01035-19. doi: 10.1128/JVI.01035-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31826999 Free PMC article.
-
Antigenic drift and subtype interference shape A(H3N2) epidemic dynamics in the United States.Elife. 2024 Sep 25;13:RP91849. doi: 10.7554/eLife.91849. Elife. 2024. PMID: 39319780 Free PMC article.
-
The antigenic architecture of the hemagglutinin of influenza H5N1 viruses.Mol Immunol. 2013 Dec;56(4):705-19. doi: 10.1016/j.molimm.2013.07.010. Epub 2013 Aug 7. Mol Immunol. 2013. PMID: 23933511 Review.
-
H3N2 influenza viruses in humans: Viral mechanisms, evolution, and evaluation.Hum Vaccin Immunother. 2018;14(8):1840-1847. doi: 10.1080/21645515.2018.1462639. Epub 2018 May 14. Hum Vaccin Immunother. 2018. PMID: 29641358 Free PMC article. Review.
Cited by
-
The roles of competition and mutation in shaping antigenic and genetic diversity in influenza.PLoS Pathog. 2013 Jan;9(1):e1003104. doi: 10.1371/journal.ppat.1003104. Epub 2013 Jan 3. PLoS Pathog. 2013. PMID: 23300455 Free PMC article.
-
Age-specific genetic and antigenic variations of influenza A viruses in Hong Kong, 2013-2014.Sci Rep. 2016 Jul 25;6:30260. doi: 10.1038/srep30260. Sci Rep. 2016. PMID: 27453320 Free PMC article.
-
Elicitation of Protective Antibodies against 20 Years of Future H3N2 Cocirculating Influenza Virus Variants in Ferrets Preimmune to Historical H3N2 Influenza Viruses.J Virol. 2019 Jan 17;93(3):e00946-18. doi: 10.1128/JVI.00946-18. Print 2019 Feb 1. J Virol. 2019. PMID: 30429350 Free PMC article.
-
Determination of antigenicity-altering patches on the major surface protein of human influenza A/H3N2 viruses.Virus Evol. 2016 Feb 14;2(1):vev025. doi: 10.1093/ve/vev025. eCollection 2016 Jan. Virus Evol. 2016. PMID: 27774294 Free PMC article.
-
A statistical analysis of antigenic similarity among influenza A (H3N2) viruses.Heliyon. 2021 Nov 12;7(11):e08384. doi: 10.1016/j.heliyon.2021.e08384. eCollection 2021 Nov. Heliyon. 2021. PMID: 34825090 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

