Predicting and analyzing DNA-binding domains using a systematic approach to identifying a set of informative physicochemical and biochemical properties
- PMID: 21342579
- PMCID: PMC3044304
- DOI: 10.1186/1471-2105-12-S1-S47
Predicting and analyzing DNA-binding domains using a systematic approach to identifying a set of informative physicochemical and biochemical properties
Abstract
Background: Existing methods of predicting DNA-binding proteins used valuable features of physicochemical properties to design support vector machine (SVM) based classifiers. Generally, selection of physicochemical properties and determination of their corresponding feature vectors rely mainly on known properties of binding mechanism and experience of designers. However, there exists a troublesome problem for designers that some different physicochemical properties have similar vectors of representing 20 amino acids and some closely related physicochemical properties have dissimilar vectors.
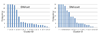
Results: This study proposes a systematic approach (named Auto-IDPCPs) to automatically identify a set of physicochemical and biochemical properties in the AAindex database to design SVM-based classifiers for predicting and analyzing DNA-binding domains/proteins. Auto-IDPCPs consists of 1) clustering 531 amino acid indices in AAindex into 20 clusters using a fuzzy c-means algorithm, 2) utilizing an efficient genetic algorithm based optimization method IBCGA to select an informative feature set of size m to represent sequences, and 3) analyzing the selected features to identify related physicochemical properties which may affect the binding mechanism of DNA-binding domains/proteins. The proposed Auto-IDPCPs identified m = 22 features of properties belonging to five clusters for predicting DNA-binding domains with a five-fold cross-validation accuracy of 87.12%, which is promising compared with the accuracy of 86.62% of the existing method PSSM-400. For predicting DNA-binding sequences, the accuracy of 75.50% was obtained using m = 28 features, where PSSM-400 has an accuracy of 74.22%. Auto-IDPCPs and PSSM-400 have accuracies of 80.73% and 82.81%, respectively, applied to an independent test data set of DNA-binding domains. Some typical physicochemical properties discovered are hydrophobicity, secondary structure, charge, solvent accessibility, polarity, flexibility, normalized Van Der Waals volume, pK (pK-C, pK-N, pK-COOH and pK-a(RCOOH)), etc.
Conclusions: The proposed approach Auto-IDPCPs would help designers to investigate informative physicochemical and biochemical properties by considering both prediction accuracy and analysis of binding mechanism simultaneously. The approach Auto-IDPCPs can be also applicable to predict and analyze other protein functions from sequences.
Figures



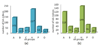

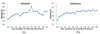


References
-
- Cai YD, Lin SL. Support vector machines for predicting rRNA-, RNA-, and DNA-binding proteins from amino acid sequence. Biochim Biophys Acta. 2003;1648(1-2):127–133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

