Chromatin higher-order structures and gene regulation
- PMID: 21342762
- PMCID: PMC3124554
- DOI: 10.1016/j.gde.2011.01.022
Chromatin higher-order structures and gene regulation
Abstract
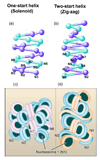
Genomic DNA in the eukaryotic nucleus is hierarchically packaged by histones into chromatin to fit inside the nucleus. The dynamics of higher-order chromatin compaction play a crucial role in transcription and other biological processes inherent to DNA. Many factors, including histone variants, histone modifications, DNA methylation, and the binding of non-histone architectural proteins regulate the structure of chromatin. Although the structure of nucleosomes, the fundamental repeating unit of chromatin, is clear, there is still much discussion on the higher-order levels of chromatin structure. In this review, we focus on the recent progress in elucidating the structure of the 30-nm chromatin fiber. We also discuss the structural plasticity/dynamics and epigenetic inheritance of higher-order chromatin and the roles of chromatin higher-order organization in eukaryotic gene regulation.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures


References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
-
-
Choy JS, Wei S, Lee JY, Tan S, Chu S, Lee TH. DNA methylation increases nucleosome compaction and rigidity. J Am Chem Soc. 2010;132:1782–1783.. ▪In this paper, the single-molecular fluorescence resonance energy transfer (FRET) method is used to show that DNA methylation increases the compaction and rigidity of nucleosomes.
-
-
- Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J Biol Chem. 2004;279:24274–24282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

