Calmodulin antagonizes a calcium-activated SCF ubiquitin E3 ligase subunit, FBXL2, to regulate surfactant homeostasis
- PMID: 21343341
- PMCID: PMC3133224
- DOI: 10.1128/MCB.00723-10
Calmodulin antagonizes a calcium-activated SCF ubiquitin E3 ligase subunit, FBXL2, to regulate surfactant homeostasis
Abstract
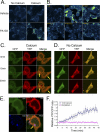
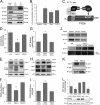
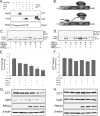
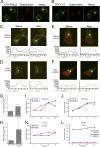
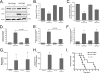
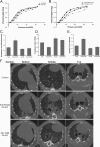
Calmodulin is a universal calcium-sensing protein that has pleiotropic effects. Here we show that calmodulin inhibits a new SCF (Skp1-Cullin-F-box) E3 ligase component, FBXL2. During Pseudomonas aeruginosa infection, SCF (FBXL2) targets the key enzyme, CCTα, for its monoubiquitination and degradation, thereby reducing synthesis of the indispensable membrane and surfactant component, phosphatidylcholine. P. aeruginosa triggers calcium influx and calcium-dependent activation of FBXL2 within the Golgi complex, where it engages CCTα. FBXL2 through its C terminus binds to the CCTα IQ motif. FBXL2 knockdown increases CCTα levels and phospholipid synthesis. The molecular interaction of FBXL2 with CCTα is opposed by calmodulin, which traffics to the Golgi complex, binds FBXL2 (residues 80 to 90) via its C terminus, and vies with the ligase for occupancy within the IQ motif. These observations were recapitulated in murine models of P. aeruginosa-induced surfactant deficiency, where calmodulin gene transfer reduced FBXL2 actions by stabilizing CCTα and lessening the severity of inflammatory lung injury. The results provide a unique model of calcium-regulated intermolecular competition between an E3 ligase subunit and an antagonist that is critically relevant to pneumonia and lipid homeostasis.
Figures


References
-
- Brandt N. R., Caswell A. H., Brandt T., Brew K., Mellgren R. L. 1992. Mapping of the calpain proteolysis products of the junctional foot protein of the skeletal muscle triad junction. J. Membr. Biol. 127:35–47 - PubMed
-
- Cenciarelli C., et al. 1999. Identification of a family of human F-box proteins. Curr. Biol. 9:1177–1179 - PubMed
-
- Chen B. B., Mallampalli R. K. 2007. Calmodulin binds and stabilizes the regulatory enzyme, CTP: phosphocholine cytidylyltransferase. J. Biol. Chem. 282:33494–33506 - PubMed
-
- Costelli P., et al. 2005. Ca2+-dependent proteolysis in muscle wasting. Int. J. Biochem. Cell Biol. 37:2134–2146 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
