The RSC chromatin remodelling enzyme has a unique role in directing the accurate positioning of nucleosomes
- PMID: 21343911
- PMCID: PMC3094113
- DOI: 10.1038/emboj.2011.43
The RSC chromatin remodelling enzyme has a unique role in directing the accurate positioning of nucleosomes
Abstract
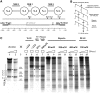
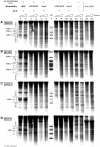
Nucleosomes impede access to DNA. Therefore, nucleosome positioning is fundamental to genome regulation. Nevertheless, the molecular nucleosome positioning mechanisms are poorly understood. This is partly because in vitro reconstitution of in vivo-like nucleosome positions from purified components is mostly lacking, barring biochemical studies. Using a yeast extract in vitro reconstitution system that generates in vivo-like nucleosome patterns at S. cerevisiae loci, we find that the RSC chromatin remodelling enzyme is necessary for nucleosome positioning. This was previously suggested by genome-wide in vivo studies and is confirmed here in vivo for individual loci. Beyond the limitations of conditional mutants, we show biochemically that RSC functions directly, can be sufficient, but mostly relies on other factors to properly position nucleosomes. Strikingly, RSC could not be replaced by either the closely related SWI/SNF or the Isw2 remodelling enzyme. Thus, we pinpoint that nucleosome positioning specifically depends on the unique properties of the RSC complex.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Basis of specificity for a conserved and promiscuous chromatin remodeling protein.Elife. 2021 Feb 12;10:e64061. doi: 10.7554/eLife.64061. Elife. 2021. PMID: 33576335 Free PMC article.
-
SWI/SNF and RSC cooperate to reposition and evict promoter nucleosomes at highly expressed genes in yeast.Genes Dev. 2018 May 1;32(9-10):695-710. doi: 10.1101/gad.312850.118. Epub 2018 May 21. Genes Dev. 2018. PMID: 29785963 Free PMC article.
-
Genome-wide nucleosome specificity and directionality of chromatin remodelers.Cell. 2012 Jun 22;149(7):1461-73. doi: 10.1016/j.cell.2012.04.036. Cell. 2012. PMID: 22726434 Free PMC article.
-
Active nucleosome positioning beyond intrinsic biophysics is revealed by in vitro reconstitution.Biochem Soc Trans. 2012 Apr;40(2):377-82. doi: 10.1042/BST20110730. Biochem Soc Trans. 2012. PMID: 22435815 Review.
-
Spanning the gap: unraveling RSC dynamics in vivo.Curr Genet. 2021 Jun;67(3):399-406. doi: 10.1007/s00294-020-01144-1. Epub 2021 Jan 23. Curr Genet. 2021. PMID: 33484328 Free PMC article. Review.
Cited by
-
Nucleosome spacing generated by ISWI and CHD1 remodelers is constant regardless of nucleosome density.Mol Cell Biol. 2015 May;35(9):1588-605. doi: 10.1128/MCB.01070-14. Epub 2015 Mar 2. Mol Cell Biol. 2015. PMID: 25733687 Free PMC article.
-
A conserved role of the RSC chromatin remodeler in the establishment of nucleosome-depleted regions.Curr Genet. 2017 May;63(2):187-193. doi: 10.1007/s00294-016-0642-y. Epub 2016 Aug 24. Curr Genet. 2017. PMID: 27558480 Free PMC article. Review.
-
Poly(dA:dT) Tracts Differentially Modulate Nucleosome Remodeling Activity of RSC and ISW1a Complexes, Exerting Tract Orientation-Dependent and -Independent Effects.Int J Mol Sci. 2023 Oct 17;24(20):15245. doi: 10.3390/ijms242015245. Int J Mol Sci. 2023. PMID: 37894925 Free PMC article.
-
The RSC chromatin remodeling complex has a crucial role in the complete remodeler set for yeast PHO5 promoter opening.Nucleic Acids Res. 2014 Apr;42(7):4270-82. doi: 10.1093/nar/gkt1395. Epub 2014 Jan 24. Nucleic Acids Res. 2014. PMID: 24465003 Free PMC article.
-
Nucleosome remodeling and epigenetics.Cold Spring Harb Perspect Biol. 2013 Sep 1;5(9):a017905. doi: 10.1101/cshperspect.a017905. Cold Spring Harb Perspect Biol. 2013. PMID: 24003213 Free PMC article. Review.
References
-
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF (2007) Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 - PubMed
-
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, Gebbia M, Talukder S, Yang A, Mnaimneh S, Terterov D, Coburn D, Li Yeo A, Yeo ZX, Clarke ND, Lieb JD et al. (2008) A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32: 878–887 - PMC - PubMed
-
- Basehoar AD, Zanton SJ, Pugh BF (2004) Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

