NeuroMap: A Spline-Based Interactive Open-Source Software for Spatiotemporal Mapping of 2D and 3D MEA Data
- PMID: 21344013
- PMCID: PMC3034234
- DOI: 10.3389/fninf.2010.00119
NeuroMap: A Spline-Based Interactive Open-Source Software for Spatiotemporal Mapping of 2D and 3D MEA Data
Abstract
A major characteristic of neural networks is the complexity of their organization at various spatial scales, from microscopic local circuits to macroscopic brain-scale areas. Understanding how neural information is processed thus entails the ability to study them at multiple scales simultaneously. This is made possible using microelectrodes array (MEA) technology. Indeed, high-density MEAs provide large-scale coverage (several square millimeters) of whole neural structures combined with microscopic resolution (about 50 μm) of unit activity. Yet, current options for spatiotemporal representation of MEA-collected data remain limited. Here we present NeuroMap, a new interactive Matlab-based software for spatiotemporal mapping of MEA data. NeuroMap uses thin plate spline interpolation, which provides several assets with respect to conventional mapping methods used currently. First, any MEA design can be considered, including 2D or 3D, regular or irregular, arrangements of electrodes. Second, spline interpolation allows the estimation of activity across the tissue with local extrema not necessarily at recording sites. Finally, this interpolation approach provides a straightforward analytical estimation of the spatial Laplacian for better current sources localization. In this software, coregistration of 2D MEA data on the anatomy of the neural tissue is made possible by fine matching of anatomical data with electrode positions using rigid-deformation-based correction of anatomical pictures. Overall, NeuroMap provides substantial material for detailed spatiotemporal analysis of MEA data. The package is distributed under GNU General Public License and available at http://sites.google.com/site/neuromapsoftware.
Keywords: CSD; Laplacian; mapping; microelectrode array; spline interpolation.
Figures
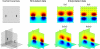

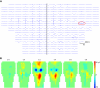



References
-
- Babiloni F., Babiloni C., Carducci F., Fattorini L., Onorati P., Urbano A. (1996). Spline Laplacian estimate of EEG potentials over a realistic magnetic resonance-constructed scalp surface model. Electroencephalogr. Clin. Neurophysiol. 98, 363–373 - PubMed
-
- Babiloni F., Babiloni C., Locche L., Cincotti F., Rossini P. M., Carducci F. (2000). High-resolution electro-encephalogram: source estimates of Laplacian-transformed somatosensory-evoked potentials using a realistic subject head model constructed from magnetic resonance images. Med. Biol. Eng. Comput. 38, 512–51910.1007/BF02345746 - DOI - PubMed
-
- Charvet G., Rousseau L., Billoint O., Gharbi S., Rostaing J. P., Joucla S., Trevisiol M., Bourgerette A., Chauvet P., Moulin C., Goy F., Mercier B., Colin M., Spirkovitch S., Fanet H., Meyrand P., Guillemaud R., Yvert B. (2010). BioMEA (TM): a versatile high-density 3D microelectrode array system using integrated electronics. Biosens. Bioelectron. 25, 1889–189610.1016/j.bios.2010.01.001 - DOI - PubMed
LinkOut - more resources
Full Text Sources

