Bacterial/archaeal/organellar polyadenylation
- PMID: 21344039
- PMCID: PMC3041983
- DOI: 10.1002/wrna.51
Bacterial/archaeal/organellar polyadenylation
Abstract
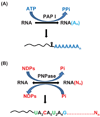
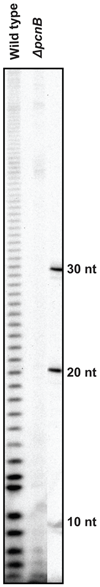
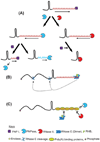
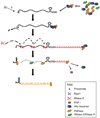
Although the first poly(A) polymerase (PAP) was discovered in Escherichia coli in 1962, the study of polyadenylation in bacteria was largely ignored for the next 30 years. However, with the identification of the structural gene for E. coli PAP I in 1992, it became possible to analyze polyadenylation using both biochemical and genetic approaches. Subsequently, it has been shown that polyadenylation plays a multifunctional role in prokaryotic RNA metabolism. Although the bulk of our current understanding of prokaryotic polyadenylation comes from studies on E. coli, recent limited experiments with Cyanobacteria, organelles, and Archaea have widened our view on the diversity, complexity, and universality of the polyadenylation process. For example, the identification of polynucleotide phosphorylase (PNPase), a reversible phosphorolytic enzyme that is highly conserved in bacteria, as an additional PAP in E. coli caught everyone by surprise. In fact, PNPase has now been shown to be the source of post-transcriptional RNA modifications in a wide range of cells of prokaryotic origin including those that lack a eubacterial PAP homolog. Accordingly, the past few years have witnessed increased interest in the mechanism and role of post-transcriptional modifications in all species of prokaryotic origin. However, the fact that many of the poly(A) tails are very short and unstable as well as the presence of polynucleotide tails has posed significant technical challenges to the scientific community trying to unravel the mystery of polyadenylation in prokaryotes. This review discusses the current state of knowledge regarding polyadenylation and its functions in bacteria, organelles, and Archaea.
Keywords: Hfq; RNA degradation; poly(A) polymerase; polynucleotide phosphorylase.
Figures

References
-
- August J, Ortiz PJ, Hurwitz J. Ribonucleic acid-dependent ribonucleotide incorporation. I. Purification and properties of the enzyme. J Biol Chem. 1962;237:3786–3793. - PubMed
-
- Hardy SJ, Kurland CG. The polynucleotide product of poly(A) polymerase from Escherichia coli. Biochemistry. 1966;5:3668–3676. - PubMed
-
- Edmonds M, Abrams R. Polynucleotide biosynthesis: Formation of a sequence of adenylate units from adenosine triphosphate by an enzyme from thymus nuclei. J Biol Chem. 1960;235:1142–1148. - PubMed
-
- Edmonds M, Abrams R. Nature of a polynucleotide required for polyribonucleotide formation from adenosine triphosphate with an enzyme from thymus nuclei. J Biol Chem. 1962;237:2636–2642. - PubMed
-
- Nakazato H, Venkatesan S, Edmonds M. Polyadenylic acid sequences in E. coli messenger RNA. Nature. 1975;256:144–146. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

